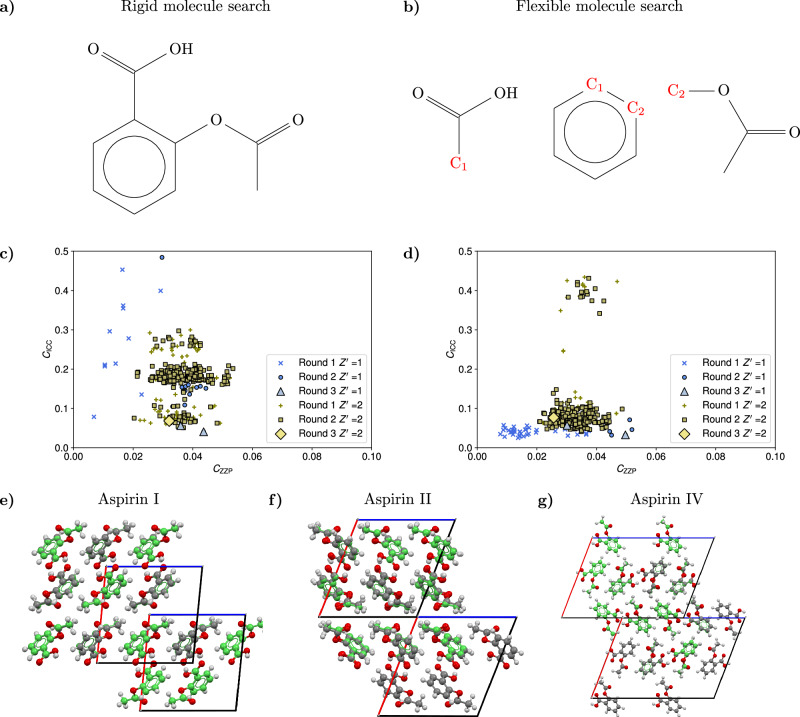
Fig. 3. Details of the crystal structure search for the aspirin molecule.
a Diagram of the complete aspirin molecule used in the rigid molecule search. b Diagram of the three rigid fragments used for the flexible molecule search of the aspirin structures. For triplets of similar unit cell geometries, the fragments are joined at the common C atoms C1, C2 (shown in red) to generate the possible conformations. c, d Scatter plots of the cost function CICC against the cost function CZZP for the three different rounds of filtering in the search for the aspirin structures. Round 1 includes all the acceptable structures with close adherence to the zero Zernike polynomial (ZZP) positions, round 2 includes structures optimized for close contacts, and round 3 contains the final accepted structures, subject to van der Waals free volume (vdWFV) and contact length constraints. High vdWFV structures were excluded from the plot. The differences in the landscapes are explained by the fact that for a given inertial eigenvector set, the orientation of the aspirin molecule in the rigid search is different from the orientation in the fragment based approach even if the conformations are similar. In addition, the rigid search conformation is not a perfect match for any flexible conformation used in the flexible search and, as a result, the structures with a rigid conformation exhibit a different adherence to the ZZPs and different contact distributions. e, f, g Overlays of the three predicted structures with the respective experimental aspirin structures, which are displayed in green. After the application of all the topological filters, only three structures are found for each search corresponding to the known aspirin I, aspirin II, and aspirin IV polymorphs, showing the agreement in the final predictions of the two searches. Source data are provided as a Source Data file.