FIGURE 4.
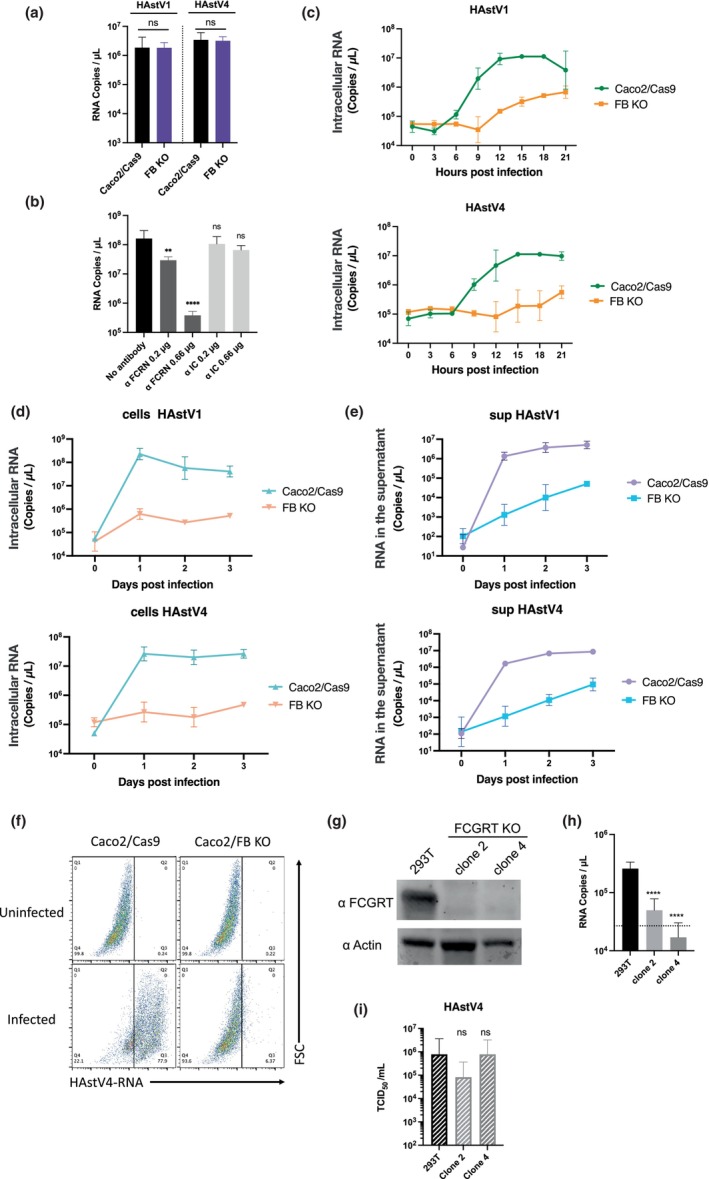
FcRn is involved in the early stages of human astrovirus replication. (a) Caco2/Cas9 cells were incubated with activated HAstV (MOI 50) at 4°C for 1 h. After washing with MEM (−) three times, RNA was extracted from the cells with the bound virus, and copies were quantified by qPCR. Each data bar represents the geometric mean of four wells. Error bars denote SD. Significance was determined by the Mann–Whitney test (ns, not significant). (b) Anti‐FCRN antibody blocked HAstV1 infection. Caco2 cells were cultured with 0.66 or 0.2 μg/ml of anti‐FCRN antibody (NBP1‐89128: Novus) or 2, 0.66, or 0.2 μg/ml rabbit isotype control IgG (ab37415: Abcam) for 1 h before infection. The Caco2 cells were incubated with the activated virus (MOI 0.005) and anti‐FCRN at the indicated concentrations for 1 h. The cells were washed three times with fresh medium and cultured for 3 days with each antibody at the indicated concentrations. Each data point represents the geometric mean of six wells. Significance was determined by Dunn's multiple comparisons test (****p < 0.0001; **p < 0.005; ns, not significant). (c) Trypsin‐activated HAstV1 or HAstV4 was inoculated into the cells (MOI 1). At each time point, the culture medium was aspirated from the wells, and the cells were washed twice with PBS. RNA was extracted, and copies were quantified by qPCR. Each point represents the mean of five wells. Error bars denote SD. (d, e) Viral RNA copies in cells and culture supernatant were reduced in FcRn‐KO cells. Trypsin‐activated HAstV1 or −4 was inoculated into the cells (MOI 1). Each day, culture medium (c) and cells (d) were collected separately. The cells were washed with PBS twice, and RNA was extracted. RNA copies in the culture medium and cells were quantified by qPCR. Each data point represents the mean of four wells. Error bars denote SD. (f) Infected cells were detected using PrimeFlow technology. Caco2 cells were exposed to trypsin‐activated HAstV4 (MOI 10) and incubated in a trypsin‐free medium for 24 h. Cells were detached from the wells and were treated with a HAstV‐specific probe according to the manufacturer's protocol. Probe‐hybridized cells were analyzed using a FACSMelody cell sorter (BD Biosciences) and FlowJo software (BD Biosciences). (g, h) Knocking out FCGRT prevents HAstV4 infection in 293T cells. (g) Detection of FCGRT in 293T cells and FCGRT‐KO clones. Actin was used as a loading control. (H) HAstV4 was incubated with trypsin to activate it. After activation, viruses were treated with FBS to inactivate trypsin and were inoculated into cells. After 1 h incubation at 37°C and subsequent washing with the medium (twice), the cells were cultured at 37°C for 3 days. The fold increase of genomic RNA copies determined by qPCR was calculated by dividing RNA copies at 3 days from 1 day. Each data bar represents the geometric mean of four wells of infected cells. Error bars denote SD. Each experiment was performed twice, and representative data are shown in this figure. Significance was determined by Dunn's multiple comparisons test (**p < 0.005; *p < 0.05; ns, not significant). (i) Viral RNA extracted from HAstV4 was transfected into 293T cells and FCGRT‐KO clones. After 72 h, the culture supernatant was serially diluted and inoculated into Caco2/Cas9 cells. After 5 days of incubation, the median tissue culture infectious dose (TCID50) was determined. Each data bar represents the geometric mean of four wells of transfected 293T cells or FCGRT‐KO clones. Each experiment was performed three times, and representative data are shown in this figure. Error bars denote SD. Significance was determined by the Dunn's multiple.
