Figure 2:
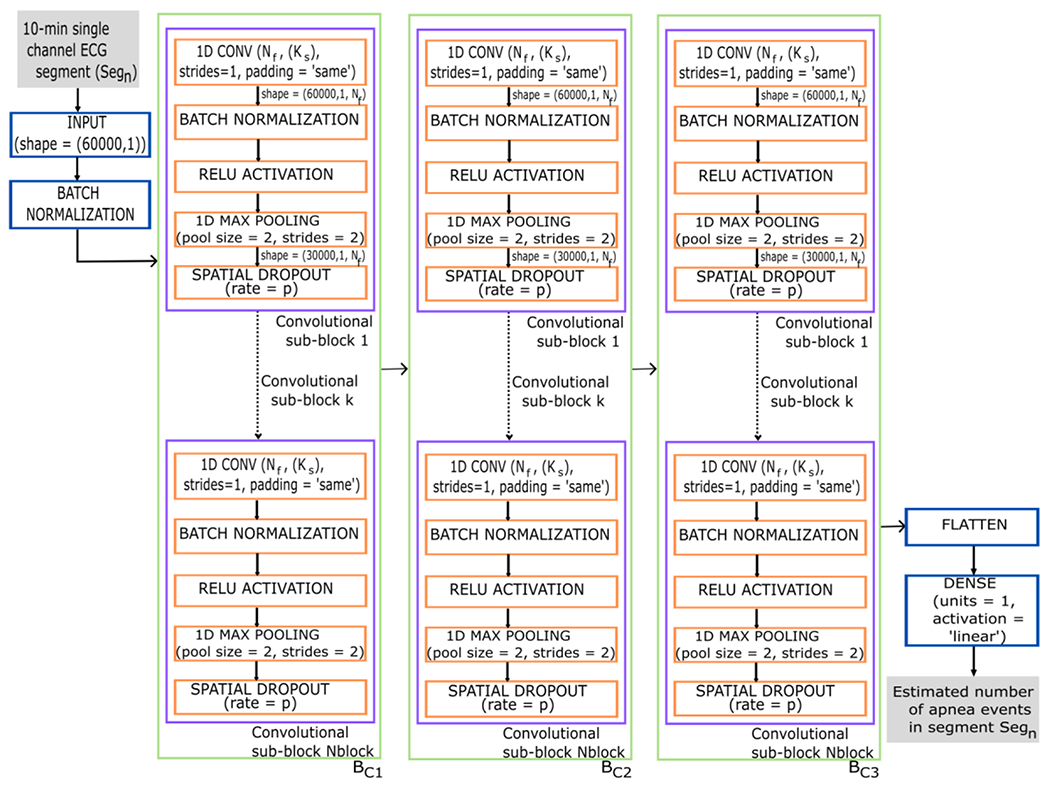
Diagram of the CNN architecture proposed in the study. The input data to the CNN are the 10-minute ECG segments and the output corresponds to the number of apnea events that the CNN estimates for each segment. 1D CONV = 1-dimensional convolutional layer; Nf = number of convolutional filters; Ks = kernel size; strides=1 in 1D CONV indicates that the stride length of the convolution is 1; padding=‘same’ in 1D CONV results in padding with zeros evenly so that the output has the same dimension as the input. RELU = rectified linear unit activation; pool size = 2 in the max pooling layer indicates that the size of the pooling window is 2; strides = 2 in the max pooling layer specifies a two-step shift of the pooling window; p = probability that each neuron is deactivated during training; Nblock = number of convolutional sub-block; BC = convolutional block 1-3. Segn is the number of the segment entering the network, ranging n from 1 to 116,126.
