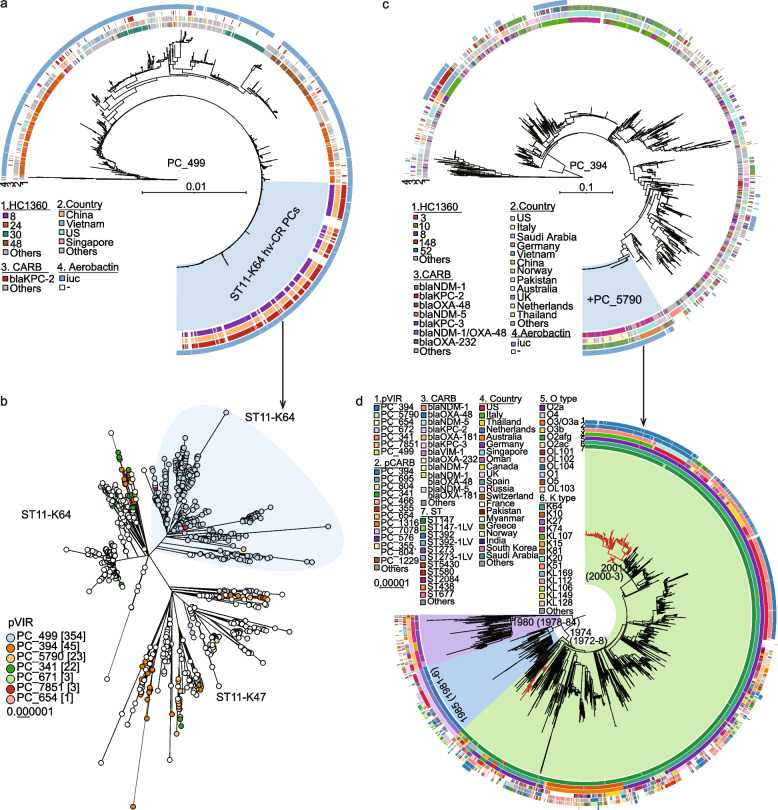
Fig. 5.
The maximum-likelihood phylogeny of the major hvCRKP HC1360 populations and their associated hvCR-PCs. a The phylogeny of 361 public PC_499 plasmids and 1141 associated plasmids predicted from assemblies. The phylogeny was built based on 7755 SNPs from 178 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 81% of the reference plasmid (AY378100). The hvCR-PCs responsible for the majority of the hvCRKP in ST11-K64 in part B are highlighted in light blue. b The phylogeny of 1114 ST11 genomes that are from China or fall within the same clade of the Chinese strains. The shape in light blue highlights a cluster of ST11-K64 strains that are mostly hvCRKPs due to the acquisition of the hvCR PC_499 plasmids in part A. c The phylogeny of 378 public PC_394 plasmids and 2608 associated plasmids predicted from assemblies. The phylogeny was built based on 28,993 SNPs from 120 KB of the conserved regions (shared by ≥ 80% of the plasmids), responsible for 40% of the reference plasmid (OW969913). The hvCR-PCs responsible for the majority of the hvCRKPs in ST147 in part d resulted from a conjugation of the plasmids in PC_5790 and are highlighted in light blue. d The phylogeny of 1493 global HC1360_3 (CC147) genomes. The three MLST STs associated with HC1360_3 (CC147) are shown in colored arcs and the hvCRKP clades are highlighted in red, carrying the hvCR PC_394 plasmids in part c. The circular bars in parts a, c, and d show metadata associated with the plasmids or genomes, as in the Keys. Visualizations of the PC_499 and PC_394 plasmid trees are available in https://itol.embl.de/shared/2Lj8mfCZAiEmU