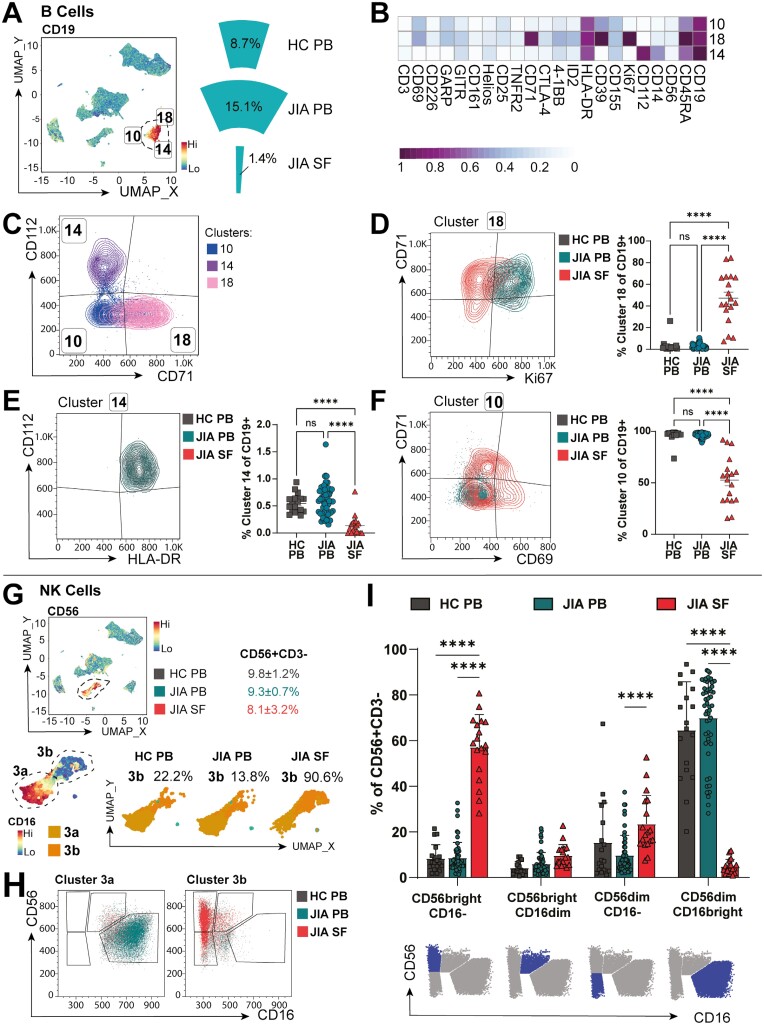
Figure 3:
JIA SF has distinct B and NK-cell populations from PB. (A) Expression UMAP of CD19 identifying three B cells clusters (10, 14, 18) with total frequencies for HC PB, JIA PB, and JIA SF (as % of total live cells) (B) Heatmap of B-cell clusters differentially expressed markers, Z-score across columns. (C) Overlay flow plot separating B-cell clusters 10, 14, and 18 by CD112 and CD71. (D–F) Left: Flow plots with right: frequencies (of % of CD19 + B cells) for clusters 18 with CD71, Ki67 (D), cluster 14 with CD112, HLA-DR (E) and cluster 10 with CD71, CD69 (F) across HC PB, JIA PB, and JIA SF. (G) Top: CD56 expression UMAP identifying NK cells with frequencies of CD56+ CD3− cells for HC PB, JIA PB, and JIA SF. Bottom: Subclassification of NK cluster (3a and 3b) based on CD16 expression with cluster 3b frequencies in HC PB, JIA PB, and JIA SF. (H) Gating strategy for NK cell subsets (cluster 3a, 3b) based on CD56 and CD16 expression with (I) frequencies of NK cell subtypes (as % of CD56+ CD3−) in HC PB, JIA PB, and JIA SF (representative gating below in blue). Throughout: JIA SF (n = 18), JIA PB (n = 52), HC PB (n = 18). Data with mean ± SEM, one/two-way ANOVA with Tukey’s multiple comparison testing, **** P < 0.0001