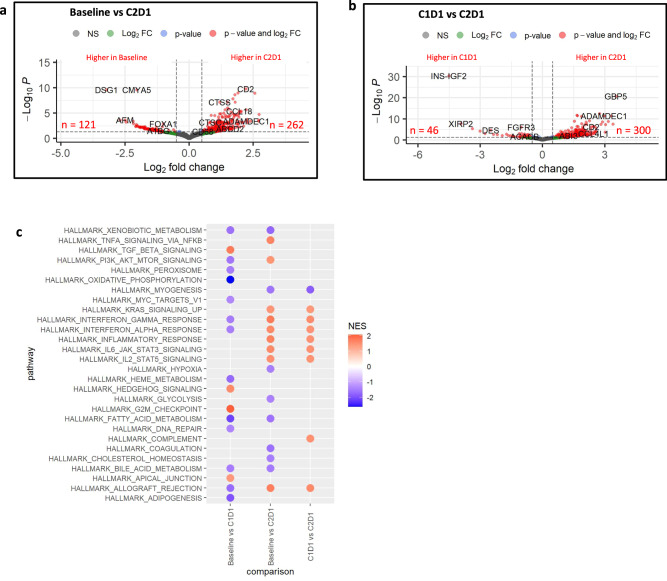
Fig. 7. RNA-seq analysis of differentially expressed genes between baseline and on treatment samples in this study cohort of PDA.
a,b Volcano plots indicating differentially expressed genes in the analysis of paired samples. The paired differential expression analysis was performed using the DESeq2 package (v1.32.0) comparing Baseline vs C2D1 samples (a) and C1D1 vs C2D1 samples (b) (n = 4 paired patient samples). Log10-transformed FDR-adjusted p-values are on the y-axis and log2-transfomed fold change between time points is on the x-axis. Genes with the absolute log2-fold changes greater than 0.5 are shown in green, genes with a FDR-adjusted p-value were below 0.05 are shown in blue, and genes that meet both thresholds are in red. c Plot of significantly differentially expressed HALLMARK pathways (FDR-adjusted p-values < 0.05) for pair-wise comparisons between time points. Gene set statistics were run with fgsea using MSigDb63 v7.4.1. Negative NES scores (blue) indicate pathways that are downregulated, while positive NES score (red) indicates pathway upregulation in C1D1 or C2D1. Source data are provided as a Source Data file. NES, normalized enrichment score.