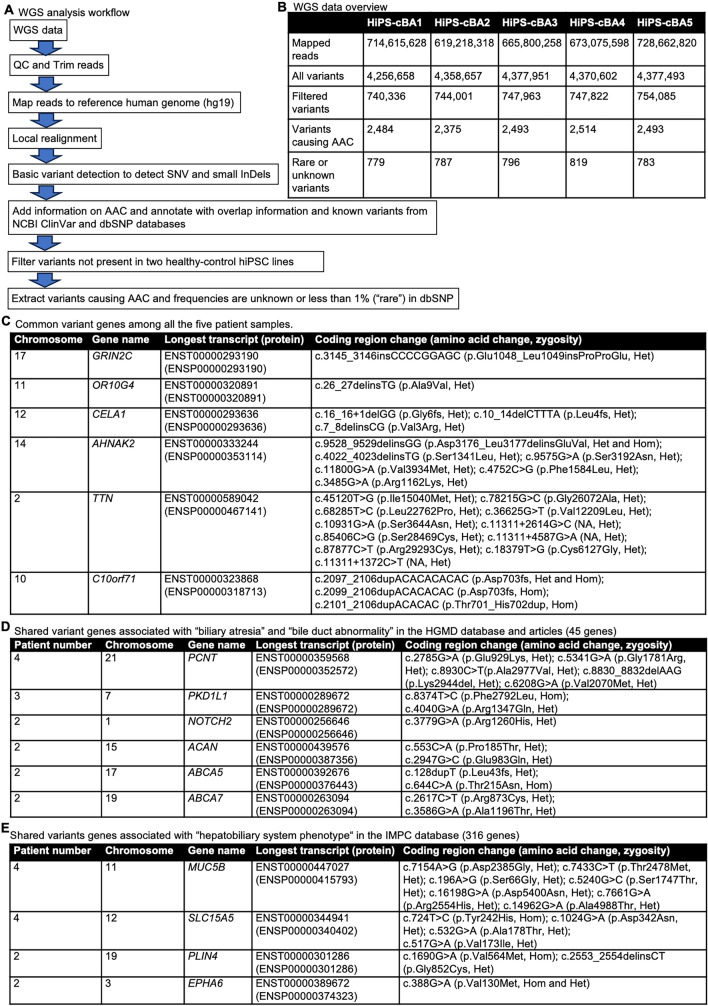
Fig. 4.
Whole genome sequencing (WGS) analysis of cystic BA patients. A Workflow of WGS analysis. QC: quality check, hg19: human genome assembly, Genome Reference Consortium (GRC) h37, SNV: single nucleotide variant, InDels: insertions or deletions, AAC: amino acid changes, and SNP: single nucleotide polymorphism. B Summary of WGS and variant numbers in the analysis of each sample. C Common variant genes among all the five patient samples. Chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown. D Shared variant genes associated with “biliary atresia” and “bile duct abnormality” in the HGMD database and articles (45 genes). Patient numbers, chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown. E Shared variant genes associated with “hepatobiliary system phenotype" in the HGMD database and articles (316 genes). Patient numbers, chromosome numbers, gene names, longest transcript and its proteins in Ensembl format, coding region change with amino acid change, and zygosity are shown