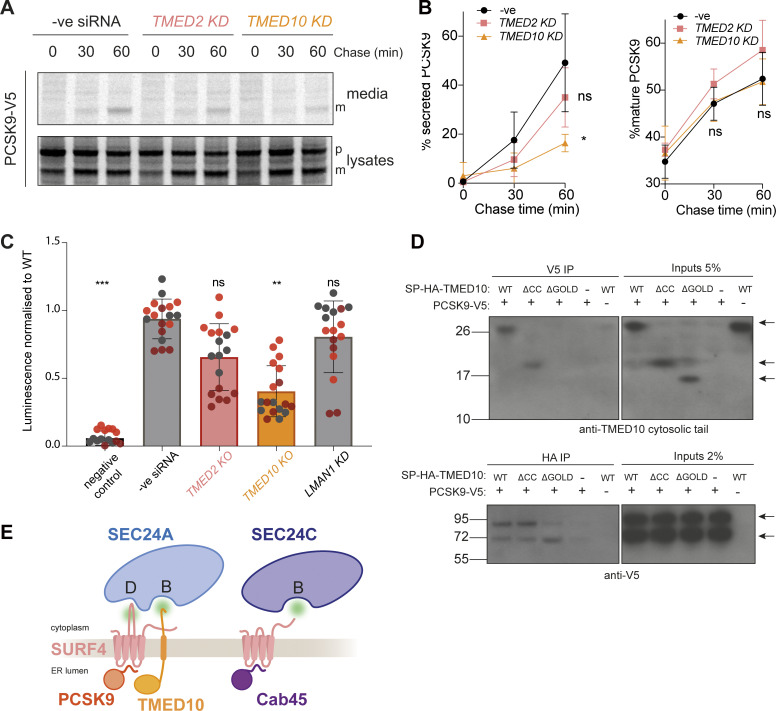
Figure 3.
TMED10 is required for efficient PCSK9 export. (A) Radiolabeled pulse-chase of PCSK9 secretion. HEK-293TREx cells were transfected with the indicated siRNAs, and after 24 h were transfected with a plasmid expressing PCSK9-V5, then subjected to pulse-chase analysis the following day. (B) Protein secretion was quantified from three biological replicates. (C) SURF4/SEC24 double KO cells were transfected with the indicated siRNAs, then after 24 h co-transfected with SmBiT-SEC24A and LgBiT-SURF4 NanoBiT constructs. Luciferase luminescence values were normalised to WT. Triangles represent mean and error bars represent SD. For each NanoBiT experiment, six technical replicates were used in each of the three independent biological replicates, as indicated by differential coloring within superplots. Triangles represent the mean and error bars represent SD. (D) DSP-crosslinking co-immunoprecipitation of PCSK9-V5 and SP-HA-TMED10 WT, coiled-coil and GOLD domain deletion mutants. The indicated constructs were co-transfected in TMED10 KO cells (- means empty pcDNA3.1 vector was used). Cells were collected, treated with DSP and cleared lysates co-immunoprecipitated overnight. Antibody to detect TMED10 signal was against TMED10 cytosolic tail. (E) A cartoon model illustrating the two mechanisms by which SURF4 relays cargo recruitment to the inner COPII coat. Statistical tests were one-way ANOVA with Dunnett’s correction for multiple testing. Data distribution was assumed to be normal but this was not formally tested. ns = not significant, * = P value <0.033, ** = P value <0.002, *** = P value <0.0002. Source data are available for this figure: SourceData F3.