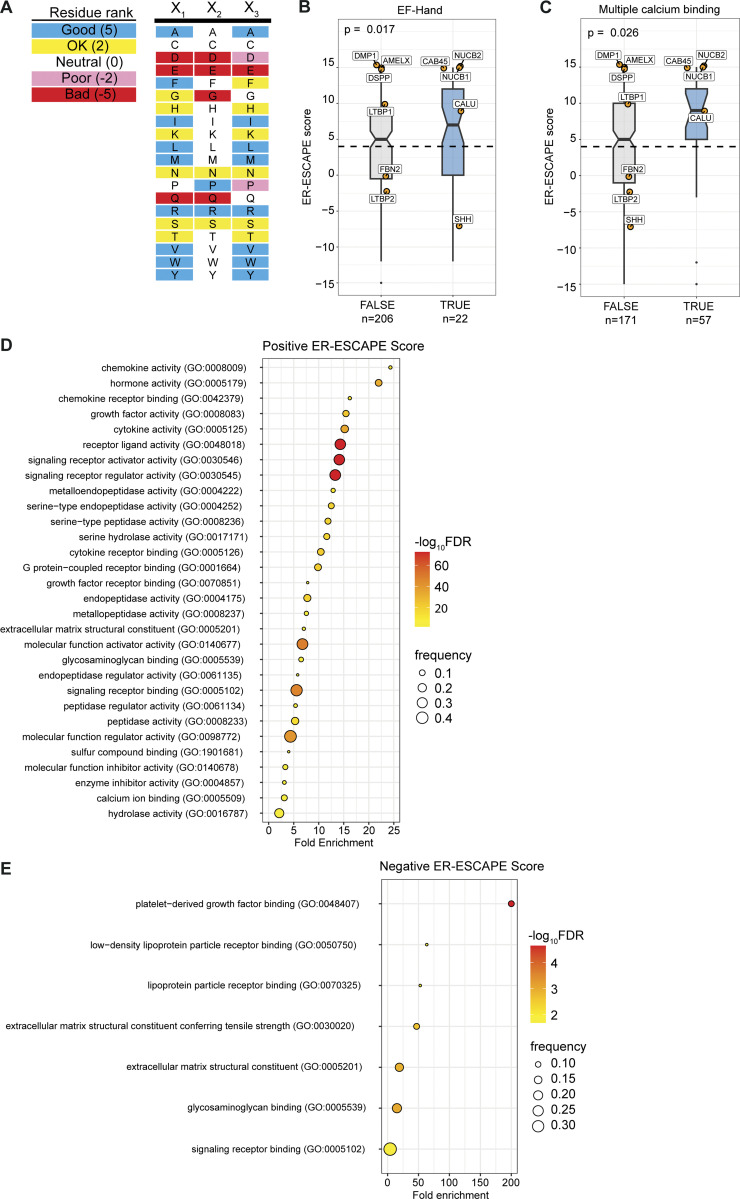
Figure S5.
ER-ESCAPE score determination and comparison of across protein properties, and Gene Ontology (GO) analysis. (A) Scoring matrix for calculating ER-ESCAPE score. (B) Among annotated calcium binding proteins in the curated soluble protein secretome, proteins with annotated EF-hand domains (PROSITE) have a significantly higher ER-ESCAPE score than those without EF-Hand annotation. (C) Among annotated calcium binding proteins in the curated soluble protein secretome, proteins with multiple calcium binding sites (according to UniProt binding site annotation) have a significantly higher ER-ESCAPE score than those with a single calcium binding site. Curated SURF4 cargoes are highlighted as orange dots with gene names. In each plot, the dashed line represents ER-ESCAPE score median of the whole dataset (n = 1988), boxes represent interquartile range, whiskers represent ranges of distribution, and notches represent 95% confidence interval of the median. Kruskal–Wallis test is used in each plot to test the significance and calculate P value. Sample size is annotated in each plot below class labels. (D and E) GO enrichment analysis for (D) high positive ER-ESCAPE score (ER-ESCAPE score = 15 or 12), or (E) negative ER-ESCAPE score (ER-ESCAPE score = −15 or −12). In each case, GO terms are sorted by fold enrichment, colored by negative logarithm of false discovery rate (FDR); the size of each dot corresponds to the frequency of observation for each term among the high-positive or negative group.