Figure 2.
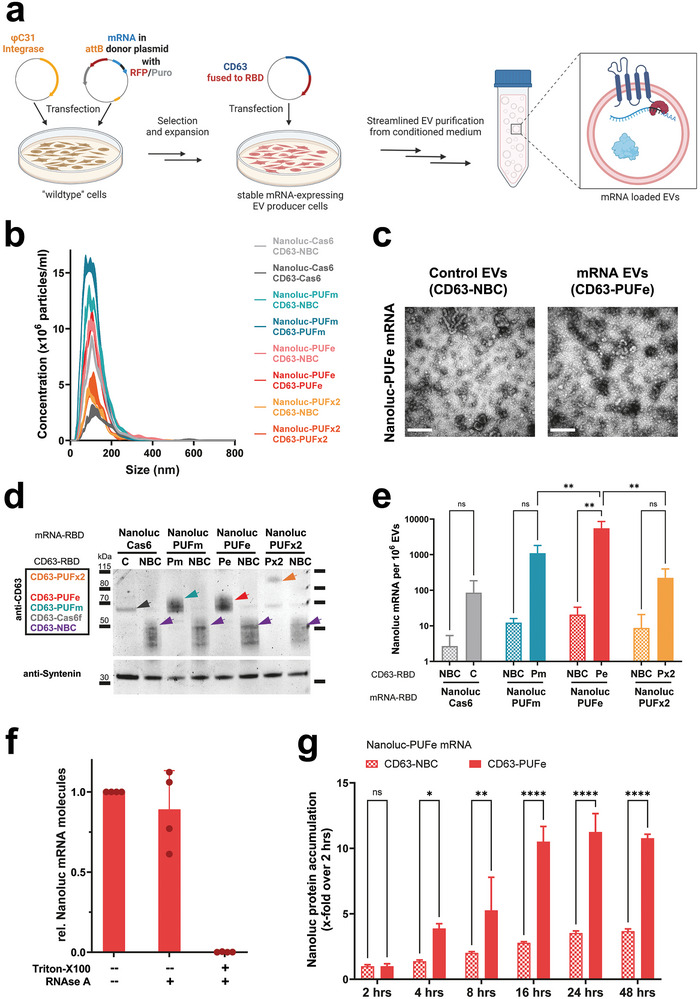
Active mRNA loading into EVs from engineered mRNA single‐stable producer cells. a) Illustration showing mRNA stable cell line generation using the ϕC31 integrase system, and subsequent EV production. For both mRNA EV and control EV production, the same mRNA stable EV producer cell line was used. Cells were either transiently expressing the compatible CD63‐RBD or incompatible CD63‐NBC (non‐binding control). This approach ensured the same biological pre‐requisites during EV biogenesis for passive loading of engineered mRNA and protein. Figure created using BioRender. b) Average particle size determination by NTA of mRNA EVs and respective control EVs harvested from transfected Nanoluc‐RBD mRNA stable producer cells (RBD motif as indicated). c) Negative stain Transmission Electron Microscopy images of control EVs and mRNA EVs loaded with Nanoluc‐PUFe mRNA. Scale bar: 300 nm d) Western Blot analysis of EVs produced from Nanoluc‐RBD mRNA stable EV producer cells as indicated on top. The expression of the CD63‐RBD fusion proteins, either CD63‐Cas6f (C), CD63‐PUFm (Pm), CD63‐PUFe (Pe), CD63‐PUFx2 (Px2), or CD63‐NBC (NBC), respectively, was validated by probing for CD63, sizes are indicated with colored arrowheads. As loading reference, expression of the EV marker SDCBP (Syntenin) was detected. Protein loaded per lane: 3 µg e) Absolute quantification by RT‐qPCR of Nanoluc mRNA molecules per 1 × 106 control EVs (NBC) or mRNA EVs (C, Pm, Pe, Px2) averaged from three representative experiments. CD63‐PUFe repeatedly showed significant enrichment of Nanoluc mRNA in EVs. Data presented as mean ± SD with n = 2 (Cas6) or n = 3 (all others); P‐values were calculated by two‐way ANOVA using Tukeys Multiple Comparisons test; α = 0.05; ns (non‐significant) p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. f) RNAse challenge assay to determine the efficiency of intraluminal EV mRNA cargo encapsulation in 4 independently produced EV batches. g) Uptake of Nanoluc mRNA EVs at a dose of 0.6 pg Nanoluc mRNA per 1 × 104 cells or particle count‐matched control EVs in Huh7 recipient cells. Cellular Nanoluc protein activity was measured at indicated timepoints and normalized to the signal measured at 2 h, which corresponds to the signal from passively loaded Nanoluc protein. Uptake of Nanoluc mRNA EVs led to a significantly higher Nanoluc protein accumulation over time compared to control EVs, demonstrating EV‐mediated engineered mRNA delivery and functional translation. Data presented as mean ± SD with n = 3; P‐values were calculated by two‐way ANOVA using Šìdàks Multiple Comparisons test; α = 0.05; ns (non‐significant) p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
