Fig. 3|. Targeting microbiota-bile acid interactions as potential therapeutic approaches for gastrointestinal and metabolic diseases.
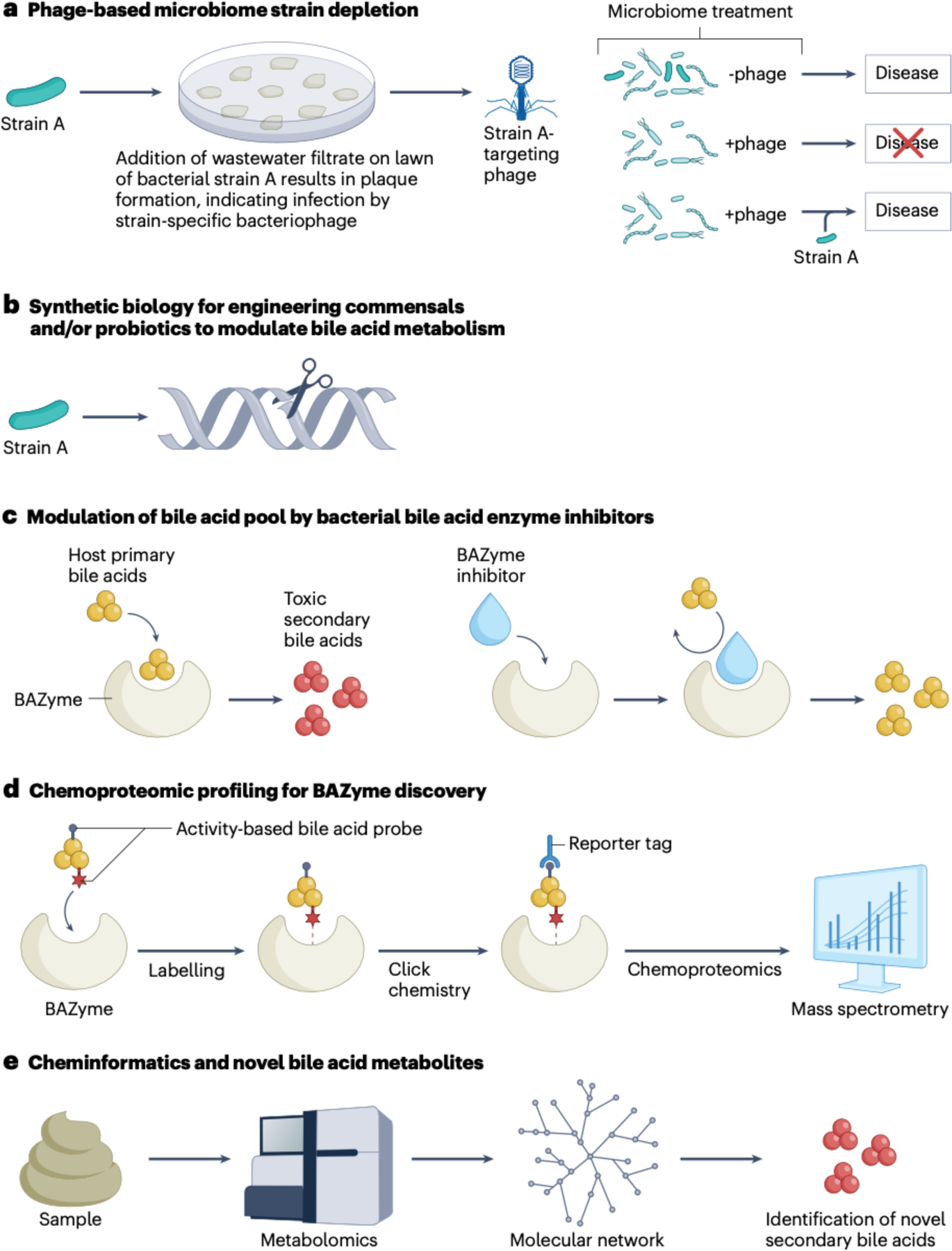
a, Studies have demonstrated the potential utility of selecting for bacterial strain-dependent bacteriophages to remove microbial strains that have a causal rolle in diseases such as inflammatory bowel disease12. b, Synthetic biology offers the potential to rationally design commensal or probiotic bacteria to modulate bile acid metabolism in vivo129. c, The development of specific inhibitors against the microblome is expected to provide therapeutic potential22. The development of bile salt hydrolase (BSH) enayme inhibitors has allowed interrogation of the effects of altering bile acid metabolism71,91,10; other studies indicate that inhibitors against bar enzymes might also be therapeutically Important11. d, Chemoproteomic profiling using click chemistry bile acid probes allows the discovery of novel bacterial enzymes involved in bile acid metabolism23. After the bile acid probe covalently bonds to a bile acid binding enzyme (BAZyme), proteomic mass spectrometry allows the identification of gene sequence candidates. e, Cheminformatics couples metabolomics with computation to obtain metabolite networks in which some nodes represent novel metabolites that repeal previously unknown bacterial metabolism121. BAZyme, bile acid enzyme.
