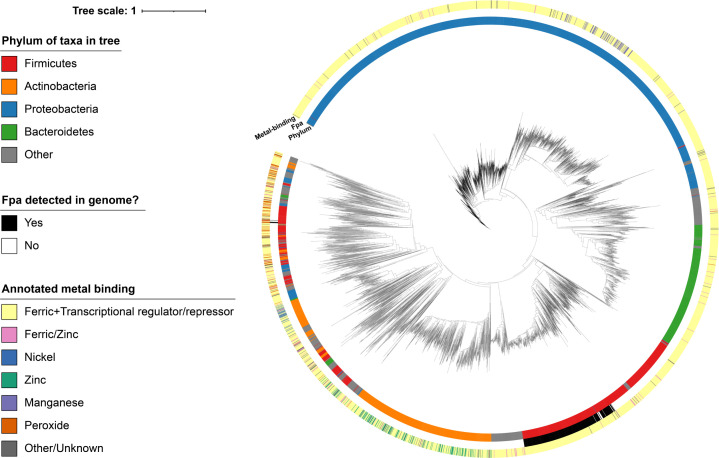
Fig 5.
Phylogeny of representative bacterial Fur sequences. Tree (midpoint rooted; built using the fastme program) of Fur protein sequences retrieved from bacterial genomes available through NCBI. Given a large number of bacterial sequences currently available through NCBI, down sampling of the retrieved Fur sequences to a genera level (i.e., only one genome per bacterial genus was considered), from just high-quality representative or reference bacterial genomes, was performed to constrain the number of sequences in the tree. For each sequence in the tree, the colors in the surrounding rings represent the phylum of the genome from which the Fur sequence was derived (inner ring), the presence or absence of F in the genome from which the Fur sequence was derived (middle ring), and annotated metal-binding affinity on NCBI of each Fur sequence (outer ring). A legend showing the colors used in the surrounding rings is shown on the left side of the image and the branch length scale.