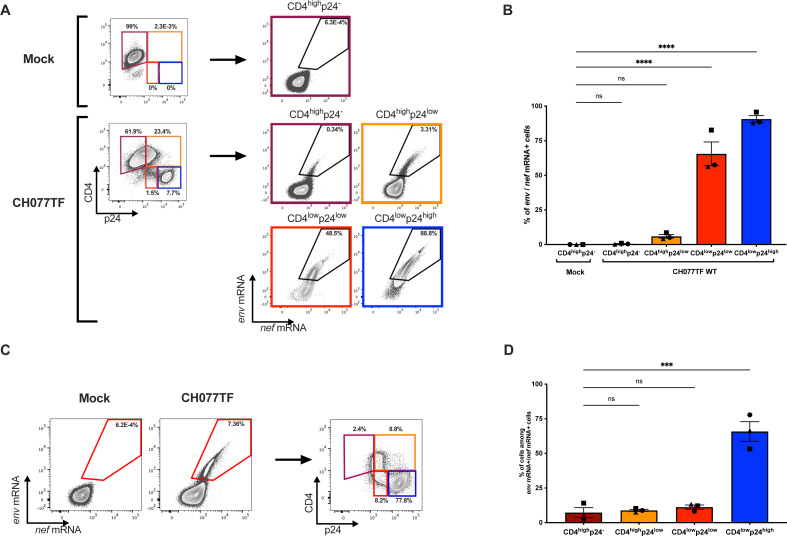
Fig 3.
HIV-1 late transcripts are mostly detected among cells that downregulated CD4. Purified primary CD4+ T cells, mock infected or infected with the transmitted-founder virus CH077 WT, were stained for cell-surface CD4 prior to detection of intracellular HIV-1 p24 and env mRNA and nef mRNA by RNA flow-FISH. (A) Representative example of flow cytometry gating strategy based on cell-surface CD4 and intracellular p24 detection and representative example of RNA flow-FISH detection of env and nef mRNA among the different cell populations. (B) Quantification of the percentage of env mRNA+ nef mRNA+ cells detected among the different cell populations in three different donors. (C) Alternatively, productively infected cells were first identified based on env and nef mRNA detection (D) Quantification of the percentage of CD4highp24−, CD4highp24low, CD4lowp24low, and CD4lowp24high cells among the env and nef mRNA+ cells with three different donors. Statistical significance was tested using one-way analysis of variance (ANOVA) with a Holm–Sidak post-test (****P < 0.0001; ns, non-significant).