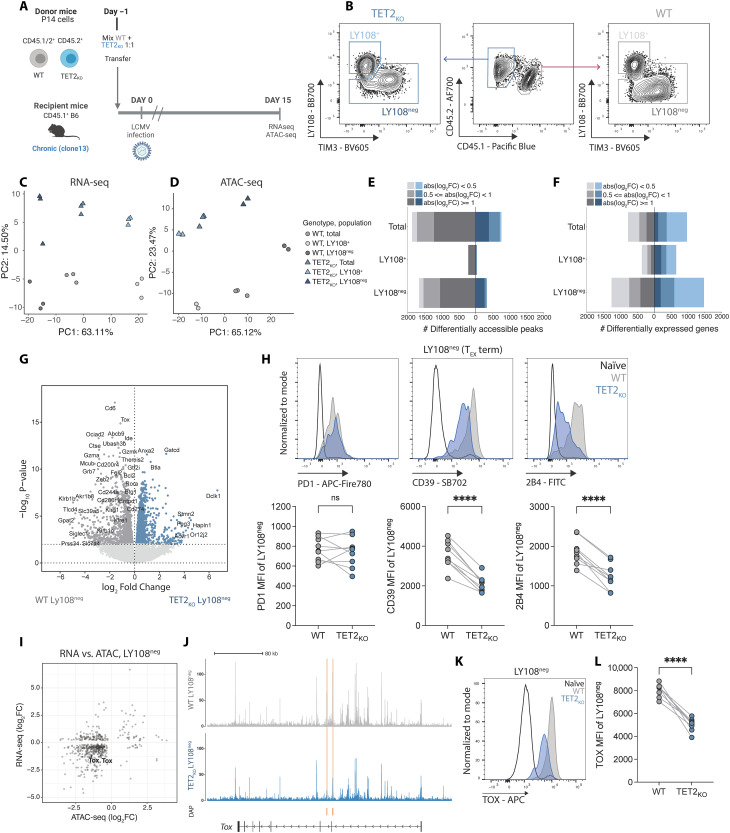
Fig. 3. Loss of TET2 limits terminal differentiation of exhausted CD8 T cells.
(A) Experiment schematic for RNA-seq and ATAC-seq. WT and TET2KO P14 cells were analyzed at day 15 p.i. with LCMV clone 13. (B) Sorting strategy for TEX subsets for RNA and ATAC-seq. (C and D) PCA of RNA-seq (C) and ATAC-seq (D) data for WT and TET2KO TEX subsets. (E and F) Number of DACRs (E) or DEGs (F) for each pairwise comparison between WT and TET2KO TEX subsets {FDR < 0.05, with variable absolute log2 fold changes [abs(log2FC)] indicated}. (G) Volcano plot highlighting DEG in WT compared to TET2KO LY108neg TEX. (H) Example plots and data comparing expression of PD1, CD39, and 2B4 on TET2KO LY108neg TEX to WT LY108neg TEX. (I) Correlation plot of differential gene expression and peak accessibility in TET2KO LY108neg TEX compared to WT LY108neg TEX with TOX labelled. (J) Example tracks showing accessibility at the Tox locus in LY108neg TEX. Differentially Accessible Peaks (DAPs) are indicated in orange. (K and L) Example plots (K) and data (L) comparing TOX expression in TET2KO LY108neg TEX to WT LY108neg TEX. [(H) and (L)] n = 9, spleen at day 30 p.i. with LCMV clone 13. Data for individual mice shown; representative of three independent experiments. ns P > 0.05; ****P < 0.0001 by paired t test. Schematic (A) created with BioRender.com.