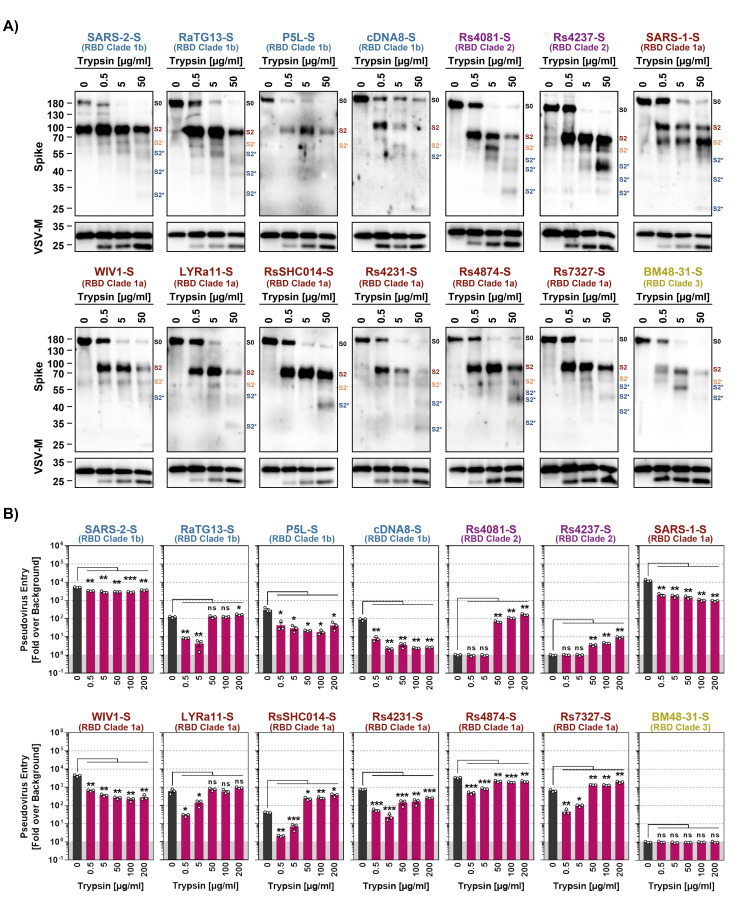
Fig 4. Trypsin cleaves the S proteins of diverse sarbecoviruses.
A) Cleavage of S proteins by trypsin. Particles pseudotyped with the indicated S proteins were incubated with the indicated concentrations of trypsin for 30 min at 37°C and S protein expression analyzed by immunoblot with SARS-CoV-2 S2 antibody. VSV-M served as loading control. Similar results were obtained in two separate experiments. Bands corresponding to uncleaved S proteins (S0), the S2 subunit (S2), S2 subunit cleaved at the S2’ site (S2’) and additional S2 cleavage fragments (S2*) are indicated and were determined based on their respective molecular weight. B) Modulation of S protein driven entry by trypsin is concentration-dependent. Particles pseudotyped with the indicated S proteins were treated with the indicated concentrations of trypsin for 30 min at 37°C before addition to Vero cells. The efficiency of S protein-driven cell entry was determined by measuring the activity of virus-encoded firefly luciferase in cell lysates at 16-18h post inoculation. Results for S protein bearing particles were normalized against those obtained for particles bearing no S protein (set as 1). The average (mean) data of three biological replicates is presented, each performed with four technical replicates. Error bars show the SEM. Statistical significance was assessed by two-tailed Student’s t-tests (p > 0.05, not significant [ns]; p ≤ 0.05, *; p ≤ 0.01, **; p ≤ 0.001, ***).