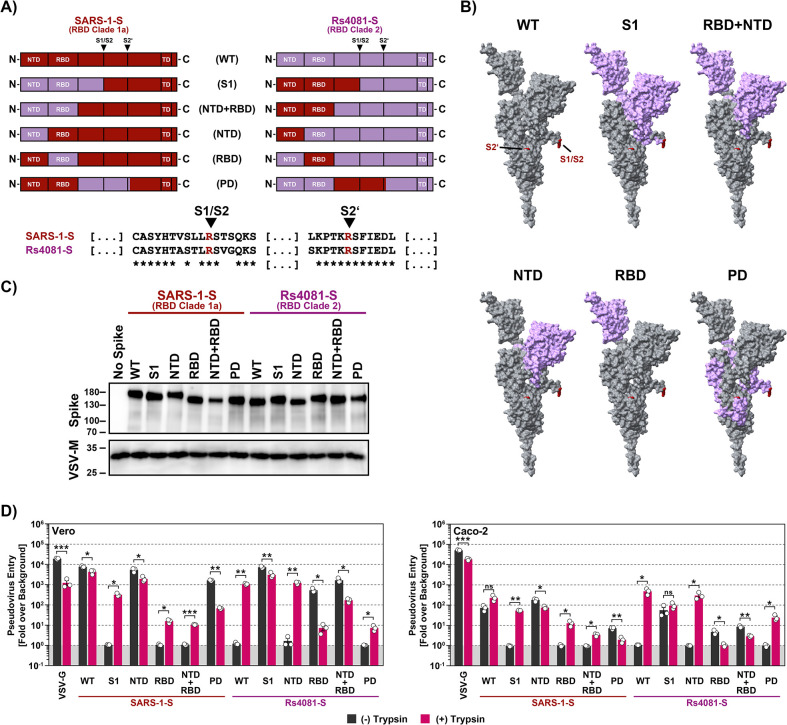
Fig 7. The RBD is the key determinant of trypsin-dependent entry.
A) Overview of the chimeric SARS-1-S and Rs4081 S proteins analyzed. The sequences of the S1/S2 and S2’ cleavage sites are indicated, asterisk indicate conserved amino acids. B) The domains exchanged between SARS-1-S and Rs4081 S proteins are color coded in the context of the S protein monomer. C) Expression of chimeric S proteins. Particles pseudotyped with the indicated S protein were subjected to immunoblot analysis, using anti an antibody directed against the S2 subunit of SARS-2-S. Detection of VSV-M served as loading control. Similar results were obtained in two separate experiments. D) Cell entry of driven by chimeric S proteins. Particles bearing the indicated S proteins (or no S protein) were treated with trypsin (50 μg/ml for 30 min at 37°C) before addition to Vero or Caco-2 cells. The efficiency of S protein-driven cell entry was determined by measuring the activity of virus-encoded firefly luciferase in cell lysates at 16-18h post inoculation. Results for S protein bearing particles were normalized against those obtained for particles bearing no S protein (set as 1). Presented are the average (mean) data of three biological replicates, each performed with four technical replicates. Error bars indicate SEM. Statistical significance was assessed by two-tailed Student’s t-tests (p > 0.05, not significant [ns]; p ≤ 0.05, *; p ≤ 0.01, **; p ≤ 0.001, ***).