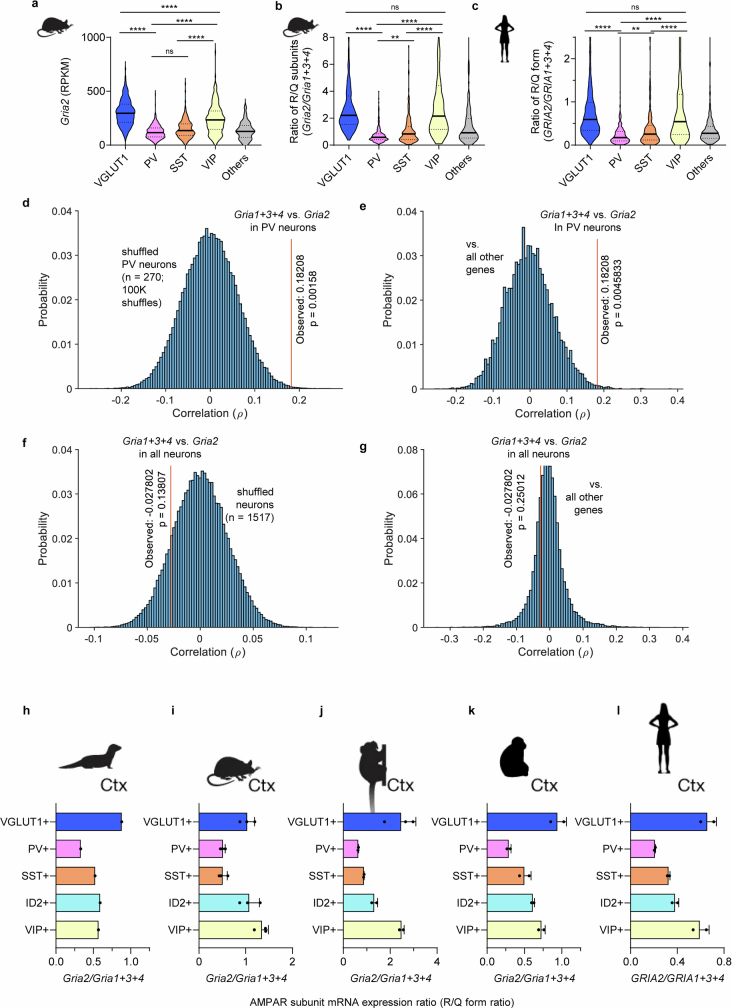
Extended Data Fig. 4. Conserved low expression of Gria2 mRNA in PV/SST interneurons across mammalian species, and potential co-regulation of Gria1-4 mRNA expression in PV interneurons.
a, Analysis of Smart-seq single-cell RNA-seq data14 from the visual cortex of p56 mice shows distinctly lower expression of Gria2 mRNA in PV and SST interneurons (n = 756/270/178/185/118 neurons from VGLUT1/PV/SST/VIP/Other cell types, respectively, = 610.9, P < 1.000x10−15, KW 1-way ANOVA; P < 0.0001 for all VGLUT1 post-hoc comparisons, Dunn’s multiple comparison correction). A fraction of outlier cells was omitted for visualization. Conventional marker protein names are adapted to denote cardinal neuronal cell classes (VGLUT1 neurons and CaMKIIα neurons both refer to forebrain excitatory neurons). Post-hoc comparisons with the ‘others’ group are omitted for brevity. b, c, This low expression of Gria2 contributes to the lower ratio of calcium impermeable/calcium permeable AMPAR subunits (R/Q subunit ratio) both in mice (b) and in humans27 (c). In both (b) and (c), a KW 1-way ANOVA test reveals a significant difference (mice: = 593.6, P < 1.000x10−15; humans: = 491.9, P < 1.000x10−15), and post-hoc comparisons demonstrate significant differences between all non-‘others’ pairs except VGLUT vs. VIP (panel c shows human data from n = 2151/235/193/282/181 neurons from VGLUT1/PV/SST/VIP/Other cell types, respectively). Post-hoc comparisons with the ‘others’ group are omitted for brevity. Thick center lines and dotted lines in violin plots represent median and 25–75% interquartile range, respectively. d-g, Potential co-regulation of Gria1-4 mRNA expression in PV interneurons. d, Single-cell mRNA expression14 of Gria2 in PV neurons showed a strong correlation (ρ = 0.18208, n = 270 cells) with the sum of Gria1, Gria3, Gria4 mRNA expression, which was highly significant compared to a bootstrap randomized distribution (100,000 shuffles across PV neurons, P = 0.0019). The Monte Carlo P-value was determined by comparing the observed correlation statistic to the simulated distribution. e, The correlation between Gria2 expression and the sum of Gria1, Gria3, Gria4 expression in PV neurons was also highly significant compared to the distribution of correlations of Gria1 + 2 + 3 with all other genes (top 0.45 percentile, P = 0.0046). f, g, This correlation was not present in the entire neuron population (n = 1517 cells, ρ = −0.027802; P = 0.1381 in comparison to shuffled neuron data; P = 0.25012 in comparison to the correlation of Gria1 + 2 + 3 to all other genes beyond Gria2). These results suggest a tight co-regulation of Gria2 vs. Gria1 + 3 + 4 mRNA expression ratio unique to PV interneurons. h-l, Conserved low expression of Gria2 mRNA in PV/SST interneurons across mammalian species. Analysis of Drop-seq single-cell RNA-seq data28 from the cortex of (h) ferrets (n = 1 replicates), (i) mice (n = 3 replicates), (j) marmosets (n = 3/2/2/2/2 replicates), (k) macaques (n = 2 replicates), and (l) humans (n = 2 replicates) shows a conserved lower ratio of calcium impermeable/calcium permeable AMPAR subunits (R/Q form ratio) in PV and SST interneurons. VGLUT1 cells correspond to cortical excitatory neurons (CaMKIIα), and ID2 correspond to neurogliaform cells. Bars and error bars denote mean ± SD of Drop-seq samples, which were averaged within replicates.