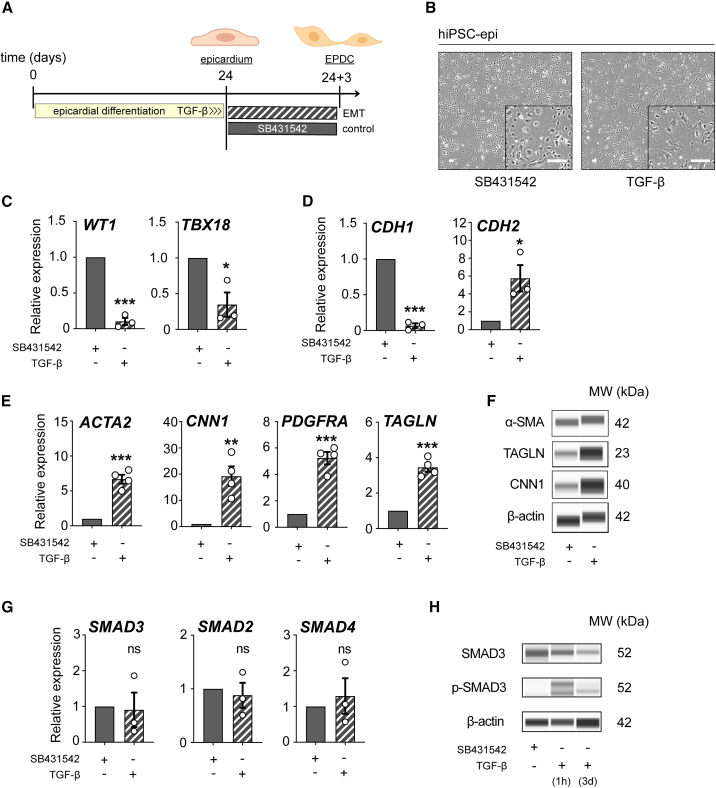
Figure 2.
SMAD3 expression dynamics during EMT in hiPSC-derived epicardial monolayers
(A) Description of the experimental workflow for epithelial-to-mesenchymal transition (EMT) induction using TGF-β and the control agent SB431542.
(B) Observations of morphological differences in hiPSC-derived EPI cells treated with TGF-β and SB431542 for 3 days, post day 24.
(C and D) Comparative qRT-PCR analysis of epicardial markers (WT1 and TBX18) (C) and genes CDH1 and CDH2 (D) in cells treated with TGF-β versus SB431542 treatment (n = 3; ∗p < 0.05, ∗∗∗p < 0.001).
(E) qRT-PCR analysis comparing the expression of epicardial-derived cell (EPDC) markers (ACTA2, CNN1, PDGFRA, and TAGLN) between TGF-β and SB431542 treatments (n = 4; ∗∗p = 0.0028, ∗∗∗p < 0.001).
(F) Western blot (Wes) analysis of EPDC markers (α-SMA, CNN1, and TAGLN) comparing TGF-β and SB431542 treatments.
(G) Comparison of qRT-PCR results for SMAD3, SMAD2, and SMAD4 between cells treated with TGF-β and SB431542 (n = 3; ns, not significant).
(H) Western blot analysis of total and phosphorylated SMAD3 in cells treated with TGF-β versus SB431542 (TGF-β treatment for 1 h is used as a positive control). All error bars represent SEM; the graph plots are derived from experimental replicates, obtained from independent batches; statistical analysis was performed using a two-sided unpaired Student’s t test; scale bars equal 100 μm, unless otherwise indicated; see also Figure S2.