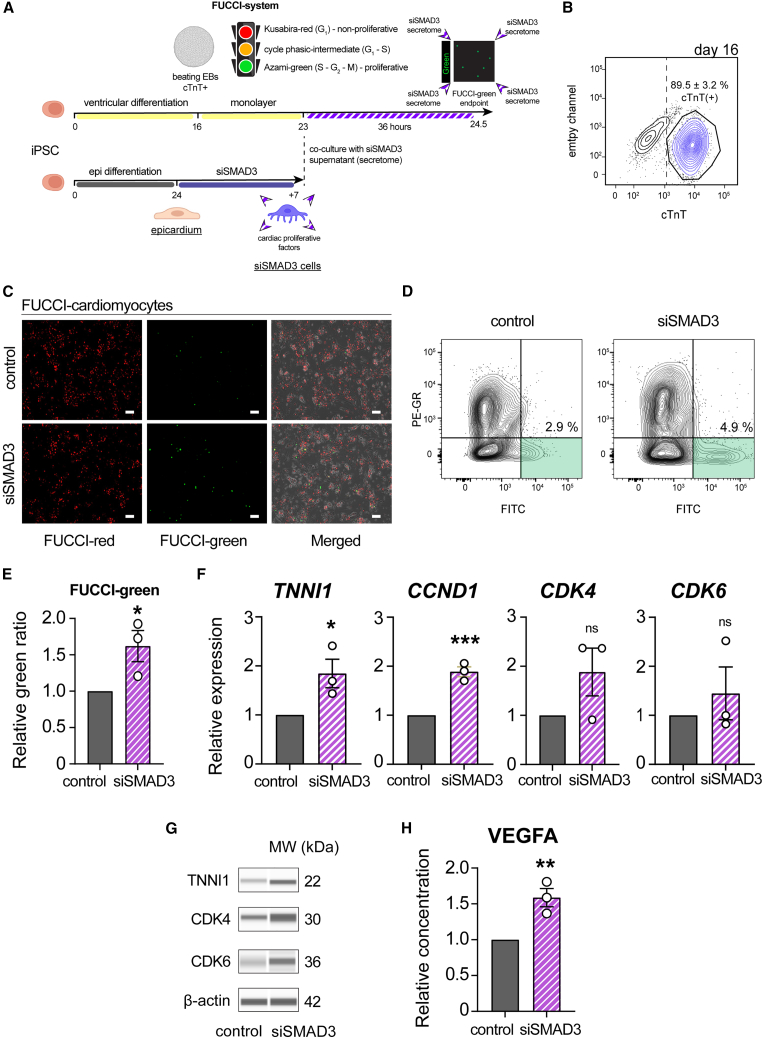
Figure 6.
Characterization of siSMAD3-derived secretome in hiPSC-CM proliferation
(A) Schematic illustration of the co-culture experiment, involving the treatment of hiPSC-derived cardiomyocytes (hiPSC-CMs) with the secretome derived from siSMAD3 treatment, to monitor cardiomyocyte proliferation using FUCCI (fluorescent ubiquitination-based cell cycle indicator) cells.
(B) Flow cytometry analysis to identify cTnT-positive cells in hiPSC-CMs at differentiation day 16 (n = 3; mean ± SEM).
(C) Fluorescence microscopy to observe FUCCI-red (non-proliferative) and FUCCI-green (proliferative) hiPSC-CMs.
(D) Representative flow cytometry contour plots analyzing FUCCI-green percentage.
(E) FUCCI-green percentage in hiPSC-CMs following treatment with the siSMAD3-derived supernatant compared to control (n = 3; ∗p = 0.044).
(F) qRT-PCR analysis for TNNI1, CCND1, CDK4, and CDK6 gene expression in hiPSC-CMs following treatment with the siSMAD3-derived supernatant (n = 3; ∗p = 0.044, ∗∗∗p = 0.0010, ns = not significant).
(G) Protein analysis in treated hiPSC-CMs for the detection of TNNI1, CDK4, and CDK6 proteins.
(H) ELISA showing that VEGFA concentration in the supernatant from SMAD3-silenced EPI cells was 49.5 pg/mL, 1.59 times compared to control (n = 3; ∗∗p = 0.010). All error bars represent SEM; the graph plots are derived from experimental replicates, obtained from independent batches; statistical analysis was performed using a two-sided unpaired Student’s t test; scale bars equal 100 μm, unless otherwise indicated; scrambled siRNA was used as a control; see also Figure S6.