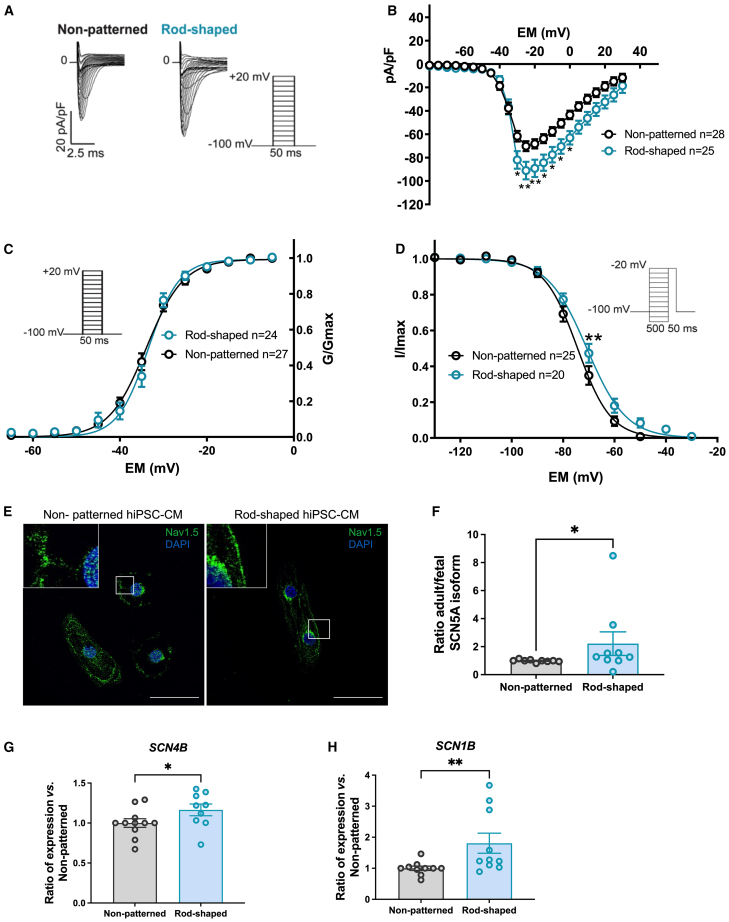
Figure 4.
Sodium channel function and expression were improved in rod-shaped hiPSC-CMs
(A) Representative INa densities in both conditions. Inset shows voltage-clamp protocol.
(B) Average peak INa densities (pA/pF) versus membrane potential (Em) in non-patterned and rod-shaped hiPSC-CMs. pA/pF, peak current amplitude to cell membrane capacitance.
(C) INa voltage dependence of activation in both conditions. GNa (as INa/(Vm−ENa), where ENa is the equilibrium potential for Na+ ions) was normalized to its maximum value (Gmax) and plotted as a function of the potential of the test pulse (inset: voltage protocol).
(D) INa voltage dependence of inactivation in both conditions. INa was normalized to its maximum value and plotted as a function of the potential of the conditioning pulse preceding the −20mV test pulse (inset: voltage protocol).
(E) Representative images showing Nav1.5 immunostaining in non-patterned and rod-shaped hiPSC-CMs at day 7 of seeding. Nuclei are shown in blue (DAPI). Scale bar, 50 μm.
(F) Ratio of adult vs. fetal SCN5A mRNA expression in non-patterned and rod-shaped hiPSC-CMs measured by SYBR green. Relative mRNA levels were normalized to non-patterned samples.
(G and H) RNA expression of SCN1B and SCN4B measured by SYBR green in non-patterned and rod-shaped hiPSC-CMs. Cts were normalized to RPL32, and the ratio of rod-shaped to non-patterned was calculated for each cardiac differentiation.
Data are expressed as mean ± SEM. For (B–D), n = 20–28 cells from 4 independent experiments. Two-way ANOVA with Bonferroni: ∗p < 0.05, ∗∗p < 0.01. For (F–H), n = 3 independent experiments from 5 to 7 differentiations per condition. Mann-Whitney, ∗p < 0.05, ∗∗p < 0.01.