ABSTRACT
Background
Recent advances in cancer genome analysis and the practice of precision medicine have made it possible to identify fractions with rare genetic alterations. Among biliary tract cancers, EGFR‐amplified cancers are known to be rare fractions across organs and have a poor prognosis. The use of anti‐EGFR antibody for EGFR‐amplified cancers has been promising; however, the evidence is not yet clear.
Case
In this report, we describe the case of a 48‐year‐old man diagnosed with advanced gallbladder cancer. The patient was administered gemcitabine plus cisplatin, followed by S‐1 monotherapy; however, disease progression was observed after two cycles of each regimen. Comprehensive genomic profiling test revealed EGFR‐amplification, and the patient was treated with combination therapy with the anti‐EGFR antibody necitumumab, gemcitabine, and cisplatin. After two cycles of treatment, tumor size reduced, and the treatment response was evaluated as partial response. On Day 90, after five cycles of treatment, tumor progression was confirmed. In addition, after disease progression, liquid biopsy revealed acquired pathogenic gene alterations suggesting anti‐EGFR antibody resistance.
Conclusion
This report supports the clinical benefit of anti‐EGFR antibodies for EGFR‐amplified biliary tract cancers and the importance of genomic analysis in personalized therapy and drug resistance research.
Keywords: biliary tract cancer, case report, drug resistance, EGFR, liquid biopsy
1. Introduction
Biliary tract cancer (BTC) is a relatively rare cancer, with an estimated incidence of fewer than six cases per 100 000 population; however, its incidence is increasing worldwide. In addition, BTC is a highly refractory cancer with one of the worst prognoses. Due in part to its anatomic complexity, early diagnosis is difficult; most patients present with locally advanced or metastatic disease at the time of diagnosis [1, 2]. Genomic profiling of BTC has revealed a variety of driver genes, and the practice of precision medicine is required [3]. Although effective molecular‐targeted drugs available for human use remain limited, anti‐EGFR antibodies have been clinically applied to cancer types that originally express high levels of EGFR. Theoretically, anti‐EGFR antibodies are expected to be effective against EGFR‐amplified cancer, and there are basic and clinical studies suggesting the utility [4, 5].
Biliary tissue is reported to express relatively high levels of EGFR, and the efficacy of anti‐EGFR antibodies against BTC has been evaluated [6]. However, all clinical trials failed to demonstrate the benefit of adding anti‐EGFR antibodies to gemcitabine (GEM)‐based regimens for advanced BTC without enrichment due to EGFR amplification [7].
Here, we describe a case of advanced EGFR‐amplified BTC that responded to combination therapy with anti‐EGFR antibody necitumumab, GEM, and cisplatin (CDDP). After treatment, acquired pathogenic gene alterations in EGFR, KRAS, RET, CTNNB1, and FH were detected by liquid biopsy.
2. Case Presentation
A 48‐year‐old man presenting with a chief complaint of epigastric pain was referred to Yokohama City University Medical Center in March of 2022. After imaging tests and tumor biopsy, the patient was diagnosed with gallbladder cancer with multiple liver and lymph node metastases (Figure 1A,B). He was administered GEM (1000 mg/m2) plus CDDP (25 mg/m2), followed by S‐1 (120 mg/body) monotherapy; however, disease progression was observed after two cycles of each regimen. A comprehensive genomic profiling test (CGP) was performed on an archival biopsy sample using the NCC Oncopanel (Sysmex Corporation, Hyogo, Japan) [8]. CGP revealed the amplification of EGFR and mutation of TP53 C135fs*35 and CDKN2A R80* (Table 1, panel A). Immunohistochemical analysis showed strong staining for EGFR and phosphorylated EGFR (Figure 1C,D, Data S1). Based on CGP, the patient was treated with necitumumab, GEM, and CDDP (necitumumab + GC) as an off‐label therapy (3‐week cycle, Day1 and 8; necitumumab [800 mg] and GEM [1000 mg/m2], Day1; CDDP [25 mg/m2]). After the initiation of the treatment, liver enzyme, lactate dehydrogenase, and tumor marker levels decreased significantly (Figure 1E–G). The value of the TP53 C135fs*35 mutation allele frequency (MAF) in cell‐free DNA (cf‐DNA) was quantified by droplet digital PCR (Bio‐Rad, CA, USA), showing a marked decrease (Figure 1H, Data S1). Contrast‐enhanced CT after two cycles of necitumumab + GC treatment showed a reduction in tumor size, and the treatment response was evaluated as partial response (Figure 1I,J). During treatment, grade 1 skin rash appeared after the first course, which could be controlled with topical steroid. Grade 3 neutropenia occurred during the third cycle, and the GEM dose was reduced (800 mg/m2). On Day 90, after five cycles of necitumumab + GC treatment, tumor progression was confirmed. After necitumumab + GC treatment, FoundationOne Liquid CDx (F1LCDx; Foundation Medicine, Boston, MA, USA) [8] revealed the emergence of pathogenic gene alterations in EGFR, KRAS, RET, CTNNB1, and FH (Table 1, panel B).
FIGURE 1.
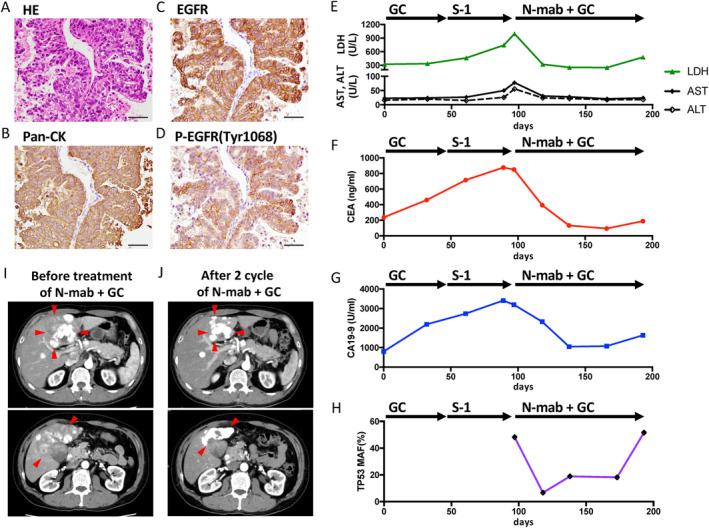
Clinical presentation. Representative pathology slides for liver biopsy specimen (A–D; scale bars represent 50 μm, 400‐fold magnification). (A) Hematoxylin and eosin, (B) pan‐cytokeratin, (C) EGFR, (D) phospho‐EGFR Tyr1068. Course of informative markers (E–H). (E) Aspartate transaminase (AST), alanine transaminase (ALT), and lactate dehydrogenase (LDH), (F) carcinoembryonic antigen (CEA), (G) carbohydrate antigen 19‐9 (CA19‐9), (H) TP53 C135fs*35 MAF in cell‐free DNA. Contrast‐enhanced CT image of the abdomen (I, J). (I) Before (baseline); (J) after 2 cycles of necitumumab + GC treatment. Arrowheads indicate the margin of the tumor. MAF, mutation allele frequency; N‐mab, necitumumab; GC, gemcitabine + cisplatin.
TABLE 1.
Summary of the detected pathogenic gene alterations.
| (A) NCC Oncopanel | |
|---|---|
| Gene | Amplification, copy number |
| EGFR | 42.60 |
| PRKCI | 12.14 |
| Gene | Mutation, amino acid replacement (mutation allele frequency [%]) |
| TP53 | C135fs*35 (68.50) |
| CDKN2A | R80* (63.30) |
| (B) FoundationOne Liquid CDx | |
|---|---|
| Gene | Amplification, copy number |
| EGFR | 6.96 |
| PRKCI | 5.03 |
| TERC | 4.63 |
| FGF10 | 2.48 |
| Gene | Mutation, amino acid replacement (mutation allele frequency [%]) |
| TP53 | C135fs*35 (48.23) |
| CDKN2A | R80* (28.37) |
| CTNNB1 | G34R (0.32), T41I (0.13), S33C (0.21), S33A (0.5), S33F (0.43), D32V (0.33), S33P (0.61), S29F (0.13), S37F (0.28), D32G (1.08), D32H (0.52) |
| EGFR | S229C (0.07), G719A (0.07) |
| KRAS | Q61H (6.62, 0.37) a |
| Gene | Rearrangement, description (mutation allele frequency [%], read count) |
| ASXL1 | ASXL1 (NM_015338) inversion intron 6–intron 11 (4.01, 52) |
| RET | CCDC6 (NM_005436)‐RET(NM_020630) fusion (C1; R12) (1.88, 91) |
| EGFR |
EGFR (NM_005228) deletion intron 1–intron 7 (EGFR vIII) (0.66, 136) EGFR (NM_005228) inversion intron 1–intron 7 (0.33, 104) EGFR (NM_005228) duplication intron 1–intron 7 (0.23, 49) EGFR (NM_005228) rearrangement intron 26 (0.23, 20) EGFR (NM_005228) deletion intron 13–intron 15 (0.14, 53) EGFR (NM_005228) rearrangement intron 25 (0.04, 12) EGFR (NM_005228) duplication intron 17–exon 27 (0.03, 61) EGFR (NM_005228) rearrangement exon 27 (0.03, 13) SEC61G (NM_014302)‐EGFR(NM_005228) fusion (S2; E13) (0.46, 206) |
| FH | FH (NM_000143) rearrangement exon 5 (5.88, 84) |
Abbreviation: EGFR vIII, EGFR variant III.
Different nucleotide variants with the same amino acid.
3. Published Data Review
To evaluate the prevalence and genomic profile of EGFR‐amplified BTC, we reviewed the previously reported tissue‐genome sequence datasets of 707 BTC samples (51 samples of cholangiocarcinoma [Firehose Legacy, TCGA], 412 samples of intrahepatic cholangiocarcinoma, and 244 samples of gallbladder cancer), available at c‐BioPortal (https://www.cbioportal.org/) [9, 10, 11, 12]. EGFR amplification was identified in 15/707 samples (2.12%), and the most common co‐existing pathogenic gene alterations included TP53 (47%) and CDKN2A (33%) (Figure 2A,B).
FIGURE 2.
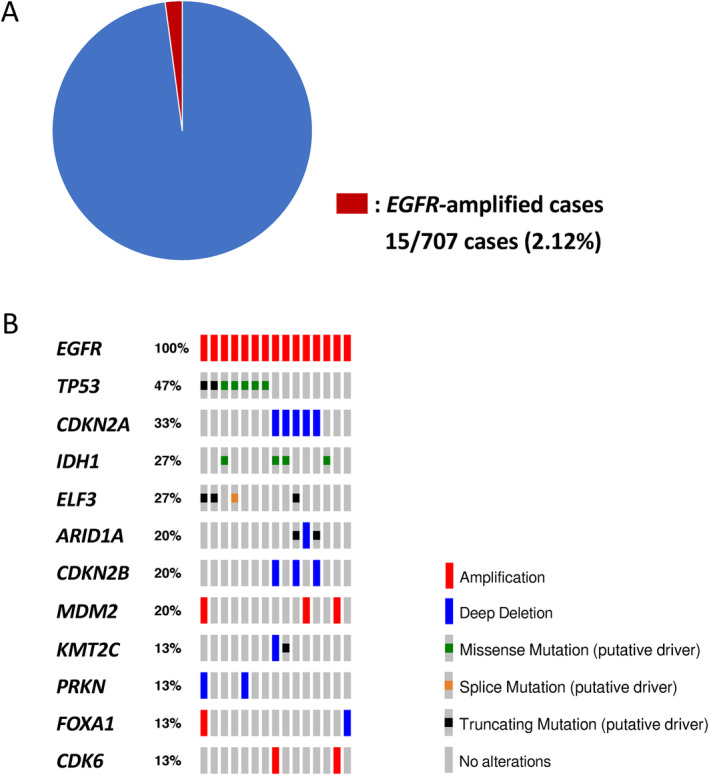
Genomic features of EGFR‐amplified biliary tract cancer in the dataset of 707 previously reported cancers, available at c‐BioPortal (https://www.cbioportal.org/) [9, 10, 11, 12]. (A) Proportion of EGFR‐amplified cases. (B) Oncoprot of 15 EGFR‐amplified cases. Gene alterations were annotated using OncoKB (https://www.oncokb.org).
4. Discussion
The recent practice of precision medicine has made it possible to identify rare fractions. We encountered a case of EGFR‐amplified BTC and reviewed 707 publicly available BTC datasets. We found that [1] EGFR‐amplified cases are extremely rare (2.12%), and [2] the coexistence of pathogenic mutations in TP53 and CDKN2A observed in the present case is a typical pattern (Table 1, Figure 2).
Of the anti‐EGFR antibodies in clinical use, necitumumab is the only IgG1 human monoclonal antibody, and it has advantages of safety and therapeutic benefits such as low risk of infusion reactions and long half‐life [13]. Further, IgG1 antibodies have high antibody‐dependent and complement‐dependent cytotoxicity, and are expected to have antitumor effects [13]. Necitumumab + GC therapy is already being clinically applied to lung cancer, [14] and we used this therapy in the present case. Tumor size reduction was observed after two cycles of necitumumab + GC treatment; hence, the effect of add‐on necitumumab to GC is evident, compared with the clinical course of the first‐line GC treatment. During the treatment, tumor markers, as well as TP53 C135fs*35 MAF values in cf‐DNA, showed a correlation with disease status. Quantitative monitoring of cf‐DNA was reported to be useful for assessing disease status, and its utility was also demonstrated [15, 16].
The timing of chemotherapy resistance is dramatic, and elucidating the mechanisms is critical. F1LCDx performed after tumor progression detected various pathogenic gene alterations (Table 1, panel B). Interestingly, numerous variants were detected in EGFR, especially in the extracellular domain (ECD), such as EGFR variant III (Table 1, panel B, Figure S1). Alterations in the ECD of EGFR are known to prevent the binding of anti‐EGFR antibodies [17, 18, 19]. Additional gene alterations detected were KRAS mutation and RET fusion, which activate RTK pathways downstream of EGFR; CTNNB1 mutation, which activates the Wnt pathway; and loss of FH, which causes oncogenic metabolic change. These findings suggest the underlying basis of anti‐EGFR antibody resistance and tumor progression. However, the limitation of this report is that CGP performed before and after treatment were not identical. Therefore, the presence of these pathogenic variants prior to treatment cannot be completely ruled out.
Anti‐EGFR antibodies have been clinically applied to colorectal cancer. Although EGFR‐amplified colorectal cancer is also rare (1%), it has been reported to be associated with a good prognosis because of the benefits of anti‐EGFR antibodies [20]. Treatment of EGFR‐amplified solid tumors remains an unmet medical need, and therapeutic strategies urgently need to be developed. To the best of our knowledge, this is the first report suggesting the therapeutic efficacy of necitumumab + GC therapy in EGFR‐amplified BTC. Based on this report, we have initiated a phase II study to investigate the efficacy of necitumumab and GEM combination therapy for EGFR‐amplified BTC (jRCTs031230259).
5. Conclusions
In this report, we describe a case of advanced EGFR‐amplified gallbladder cancer that responded to combination therapy with the anti‐EGFR antibody necitumumab, GEM, and CDDP. In addition, we used liquid biopsy to monitor the development of resistance and elucidate the molecular mechanism of resistance. A review of the published data indicates that BTCs with EGFR‐amplification are a rare fraction (accounting for about 2.1%); however, effective utilization of existing drugs is a pressing issue. Hence, treatment with anti‐EGFR antibodies for EGFR‐amplified BTCs should be validated by further studies.
Author Contributions
Makoto Sugimori: conceptualization, methodology, data curation, writing – original draft, project administration. Masaki Nishimura: conceptualization, writing – original draft, methodology, project administration, data curation. Kazuya Sugimori: conceptualization, writing – review and editing. Sho Tsuyuki: conceptualization, writing – review and editing. Akane Hirotani: conceptualization, writing – review and editing. Haruo Miwa: conceptualization, writing – review and editing. Takashi Kaneko: conceptualization, writing – review and editing. Haruka Hirose: data curation, methodology, project administration. Yoshiaki Inayama: methodology, data curation, project administration. Akito Nozaki: conceptualization, project administration, supervision, writing – review and editing. Kazushi Numata: conceptualization, writing – review and editing, project administration, supervision. Chikara Kunisaki: conceptualization, writing – review and editing, project administration, supervision. Shin Maeda: conceptualization, writing – review and editing, project administration, supervision.
Ethics Statement
The off‐label use of the drug limited to this patient was approved by the Institutional Review Board of Yokohama City University Medical Center. Analysis of the cf‐DNA was approved by the Regional Committee for Medical and Health Research Ethics of Yokohama City University (approval number: B160804006), and the patient was provided informed consent prior to the specimen collection.
Consent
Informed consent for publication of this case report has been obtained from the patient.
Conflicts of Interest
The authors declare no conflicts of interest.
Supporting information
Figure S1.
Data S1.
Acknowledgments
We thank S. Yonei, Y. Matsuoka, H. Yoshimura, K. Endo, R. Oishi, H. Tsuchiya, S. Komiyama, E. Ikeda, M. Kosugi, F. Kobayashi, H. Tanaka, T. Kodera, and A. Takase for their support in this work.
Funding: This study was funded by the Kihara Memorial Yokohama Foundation for the Advancement of Life Science (Grant No. 2022,1046‐28) and the Yokohama City University Research Grant for the Promotion of Advanced Medicine. The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Makoto Sugimori and Masaki Nishimura contributed equally to this work.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1. Banales J. M., Marin J. J. G., Lamarca A., et al., “Cholangiocarcinoma 2020: The Next Horizon in Mechanisms and Management,” Nature Reviews. Gastroenterology & Hepatology 17, no. 9 (2020): 557–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim D., Konyn P., Cholankeril G., Bonham C. A., and Ahmed A., “Trends in the Mortality of Biliary Tract Cancers Based on Their Anatomical Site in the United States From 2009 to 2018,” American Journal of Gastroenterology 116, no. 5 (2021): 1053–1062. [DOI] [PubMed] [Google Scholar]
- 3. Karasic T. B., Eads J. R., and Goyal L., “Precision Medicine and Immunotherapy Have Arrived for Cholangiocarcinoma: An Overview of Recent Approvals and Ongoing Clinical Trials,” JCO Precision Oncology 7 (2023): e2200573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakamura Y., Sasaki A., Yukami H., et al., “Emergence of Concurrent Multiple EGFR Mutations and MET Amplification in a Patient With EGFR‐Amplified Advanced Gastric Cancer Treated With Cetuximab,” JCO Precision Oncology 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kato S., Okamura R., Mareboina M., et al., “Revisiting Epidermal Growth Factor Receptor (EGFR) Amplification as a Target for Anti‐EGFR Therapy: Analysis of Cell‐Free Circulating Tumor DNA in Patients With Advanced Malignancies,” JCO Precision Oncology 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kawamoto T., Ishige K., Thomas M., et al., “Overexpression and Gene Amplification of EGFR, HER2, and HER3 in Biliary Tract Carcinomas, and the Possibility for Therapy With the HER2‐Targeting Antibody Pertuzumab,” Journal of Gastroenterology 50, no. 4 (2015): 467–479. [DOI] [PubMed] [Google Scholar]
- 7. Rizzo A., Frega G., Ricci A. D., et al., “Anti‐EGFR Monoclonal Antibodies in Advanced Biliary Tract Cancer: A Systematic Review and Meta‐Analysis,” In Vivo 34, no. 2 (2020): 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ebi H. and Bando H., “Precision Oncology and the Universal Health Coverage System in Japan,” JCO Precision Oncology 3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boerner T., Drill E., Pak L. M., et al., “Genetic Determinants of Outcome in Intrahepatic Cholangiocarcinoma,” Hepatology 74, no. 3 (2021): 1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giraldo N. A., Drill E., Satravada B. A., et al., “Comprehensive Molecular Characterization of Gallbladder Carcinoma and Potential Targets for Intervention,” Clinical Cancer Research 28, no. 24 (2022): 5359–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerami E., Gao J., Dogrusoz U., et al., “The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data,” Cancer Discovery 2, no. 5 (2012): 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao J., Aksoy B. A., Dogrusoz U., et al., “Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal,” Science Signaling 6(269):pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dienstmann R. and Tabernero J., “Necitumumab, a Fully Human IgG1 mAb Directed Against the EGFR for the Potential Treatment of Cancer,” Current Opinion in Investigational Drugs 11, no. 12 (2010): 1434–1441. [PubMed] [Google Scholar]
- 14. Thatcher N., Hirsch F. R., Luft A. V., et al., “Necitumumab Plus Gemcitabine and Cisplatin Versus Gemcitabine and Cisplatin Alone as First‐Line Therapy in Patients With Stage IV Squamous Non‐small‐Cell Lung Cancer (SQUIRE): An Open‐Label, Randomised, Controlled Phase 3 Trial,” Lancet Oncology 16, no. 7 (2015): 763–774. [DOI] [PubMed] [Google Scholar]
- 15. Sugimori M., Sugimori K., Tsuchiya H., et al., “Quantitative Monitoring of Circulating Tumor DNA in Patients With Advanced Pancreatic Cancer Undergoing Chemotherapy,” Cancer Science 111, no. 1 (2020): 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krebs M. G., Malapelle U., André F., et al., “Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients With Cancer: A Narrative Review,” JAMA Oncology 8, no. 12 (2022): 1830–1839. [DOI] [PubMed] [Google Scholar]
- 17. Gan H. K., Cvrljevic A. N., and Johns T. G., “The Epidermal Growth Factor Receptor Variant III (EGFRvIII): Where Wild Things Are Altered,” FEBS Journal 280, no. 21 (2013): 5350–5370. [DOI] [PubMed] [Google Scholar]
- 18. Chong C. R. and Jänne P. A., “The Quest to Overcome Resistance to EGFR‐Targeted Therapies in Cancer,” Nature Medicine 19, no. 11 (2013): 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. An Z., Aksoy O., Zheng T., Fan Q. W., and Weiss W. A., “Epidermal Growth Factor Receptor and EGFRvIII in Glioblastoma: Signaling Pathways and Targeted Therapies,” Oncogene 37, no. 12 (2018): 1561–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Randon G., Yaeger R., Hechtman J. F., et al., “EGFR Amplification in Metastatic Colorectal Cancer,” Journal of the National Cancer Institute 113, no. 11 (2021): 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Data S1.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


