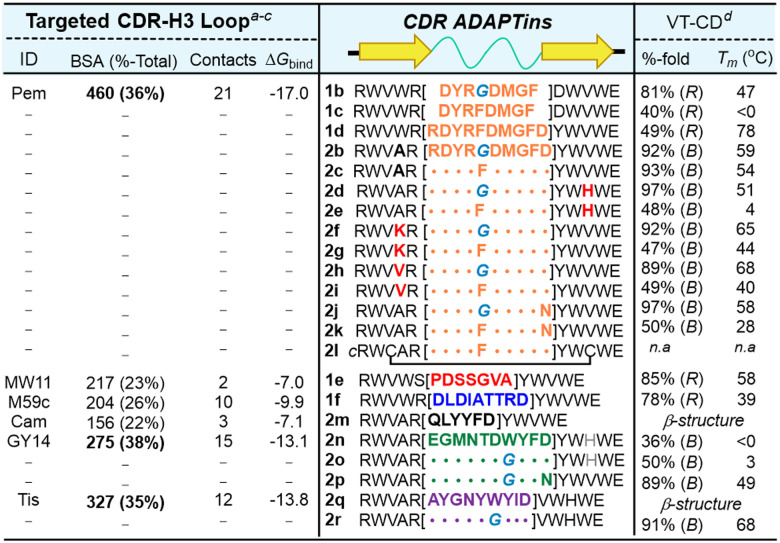
Library of ADAPTins with diversely functionalized H3 loops generated from an individual CDR-H3 binding analysis. Evaluation of folding and stability properties of the designed ADAPTin peptides 1–2.
Individual CDR buried surface area (BSA) computed by dr_SASA with %-binding surface calculated as a ratio to the total BSA (TBSA) from Ab·PD1 cocrystal structures.
Total number of hydrophobic and polar binding contacts created at the CDR-H3 interface.
Binding Gibbs free energy (reu) calculated by Peptiderive to score binding interfaces.
Folded fraction (χF) at 25 °C and melting temperatures calculated from CD-melts based on the type of ADAPTin fold (B: bulged, R: regular).