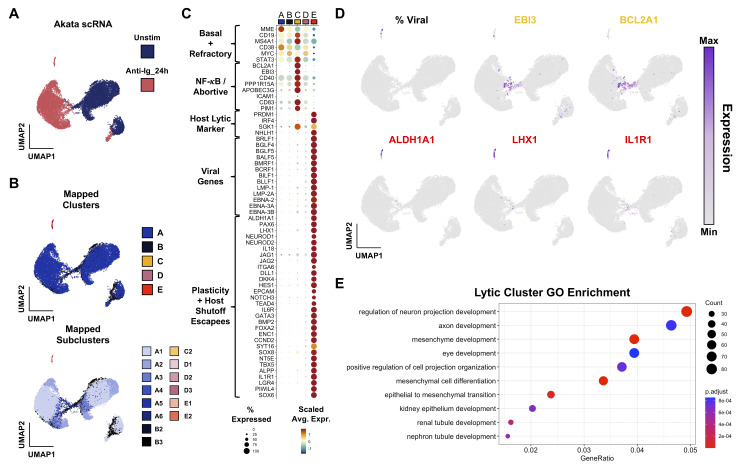
Fig 7. Validation of abortive and reprogramming gene expression signatures in Akata BL cells stimulated by anti-Ig.
(A) Integrated scRNA-seq data from Akata cells without stimulation (blue) and 24 h after anti-Ig treatment to induce lytic reactivation (rose). (B) Cluster and subcluster mapping from P3HR1-ZHT data to Akata data via transfer anchor integration. Clusters are colored as for P3HR1-ZHT and B958-ZHT datasets. Colors represent the reference phenotype with the maximal integration score for each cell. (C) Cluster-resolved expression in integrated Akata scRNA-seq dataset. (D) UMAP visualization of viral read fraction and representative markers of abortive (yellow) and reprogramming (red) signatures. LHX1 and IL1R1 are known escapees of host shutoff. (E) GO enrichment of DE genes upregulated in anti-Ig-induced Akata lytic cells.