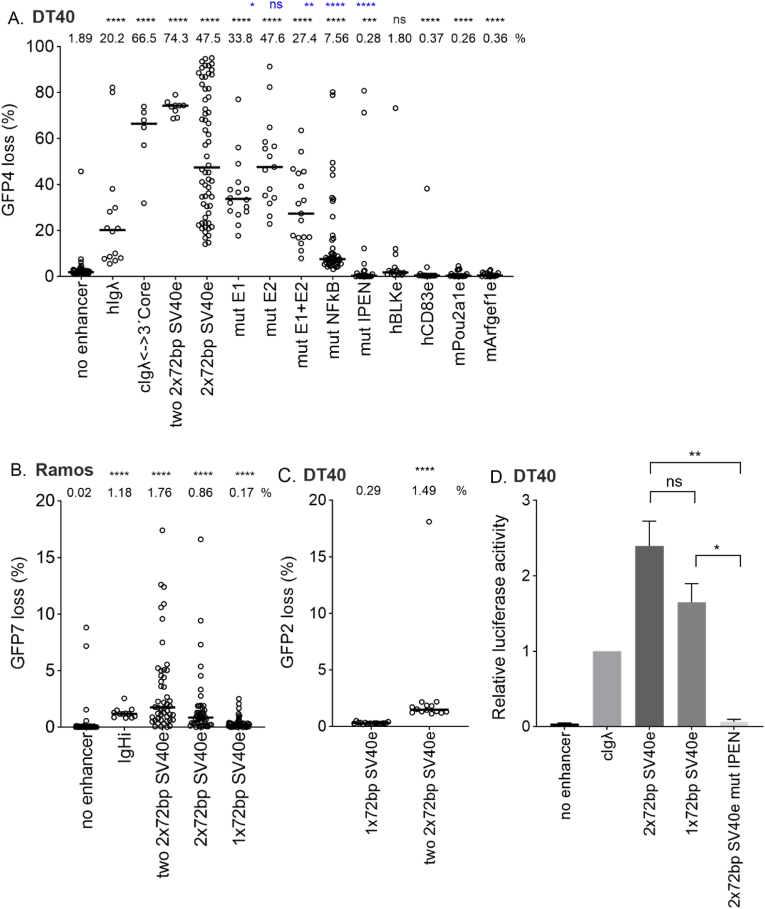
Fig. 2.
SV40 enhancer SHM targeting activity in chicken DT40 cells and human Ramos B cells.
A. SHM targeting activity of two 2x72bp SV40 enhancer, 2x72bp SV40 enhancer and its TFBS mutants as well as four genomic enhancers (human BLKe, human CD83, mouse Obf1 and mouse Cd79) measured in GFP loss assay in GFP4 reporter in chicken DT40 cell line. Empty GFP4 reporter vector was used as negative control and human Igλ (hIgλ) enhancer core and chicken Igλ enhancer 3′ core (cIgλ<−>3′core) were used as positive controls. Median GFP loss (%) values are shown above the graph. Statistical significance compared to negative control is indicated in black and statistical significance compared to 2x72bp enhancer is indicated in blue. Mann-Whitney U test was used to calculate statistical significances. Mutated TFBS binding site(s) are indicated in x-axes naming. IPEN: IRF-Ets, E-box and NF-KB binding site mutated.
B. SHM targeting activity of two 2x72bp SV40 enhancer, 2x72bp SV40 enhancer and 1x72bp SV40 enhancer in Ramos human B cell line. Empty GFP7 reporter vector was used as negative control and human Ig heavy chain intronic enhancer core (IgHi) was used as positive control. Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significances.
C. SHM targeting activity of 1x72bp SV40 enhancer and two 2x72bp SV40 enhancer measured in GFP loss assay in GFP2 reporter in chicken DT40 cell line. Median GFP loss (%) values are shown above the graph. Mann-Whitney U test was used to calculate statistical significance. The percentages of GFP loss in panels A, B and C are not directly comparable with each other due to the differences in sensitivity and host cell between GFP2, GFP4 and GFP7 reporters.
D. Enhancer activity of 2x72bp SV40 enhancer, 1x72bp SV40 enhancer and 2x72bp SV40 IPEN mutant enhancer measured in luciferase assay. Chicken Igλ enhancer (cIgλ) was used as positive control and empty vector as negative control. Two replicate measurements were performed. Fold changes over cIgλE are shown. Unpaired t-test was used to calculate statistical significances. ∗∗∗∗ p<0.0001, ∗∗∗ p<0.001, ∗∗ p<0.01, ∗ p<0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)