Fig. 4.
Mutation type and distribution of LT region in Ramos and UO-31 cells.
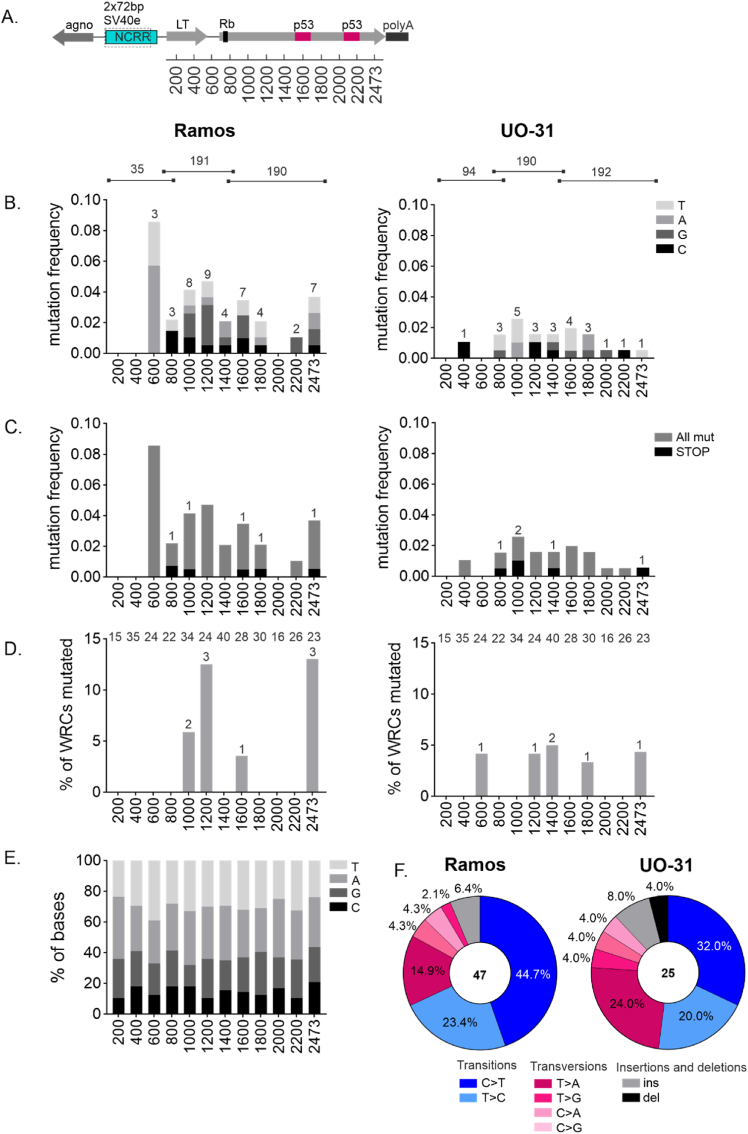
A. A diagram of genome-inserted SV40 LT transcription unit with locations of Rb1-binding site and two-part p53 binding site. Location of bins is shown below the figure
B. Overall mutation distribution along SV40 LT region. Mutations targeting different bases are indicated in different colors. Mutation frequency (number of mutations/number of sequences per bin) is shown in the y-axis. Number of mutations observed in each bin is shown. Number of sequences obtained from Ramos and UO-31 LT region is shown above the graphs and concerns panels B–D.
C. Overall and STOP-codon forming mutation distribution along SV40 LT region. Overall and STOP-mutation are indicated in different colors. Number of STOP codon forming mutations is shown. Mutation frequency (number of mutations/number of sequences per bin) is shown in the y-axis.
D. % of mutations targeting AID hotspot WRC per bin along SV40 LT. Number of mutated WRC per bin is shown. Number of WRC motifs per bin in LT region is indicated on the top of the graphs.
E. Each bin of SV40 LT presented by its base content.
F. Mutation types in LT area. Substitutions are presented in six substitution classes: C > A, C > T, C > G, T > A, T > C, T > G. Substitutions are also divided to transitions (blue) and transversions (red) and are arranged accordingly. In addition, deletions and insertions (grey and black) are shown.
For panels BE LT area is divided to 200 bp bins except final bin being 273 bp. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)