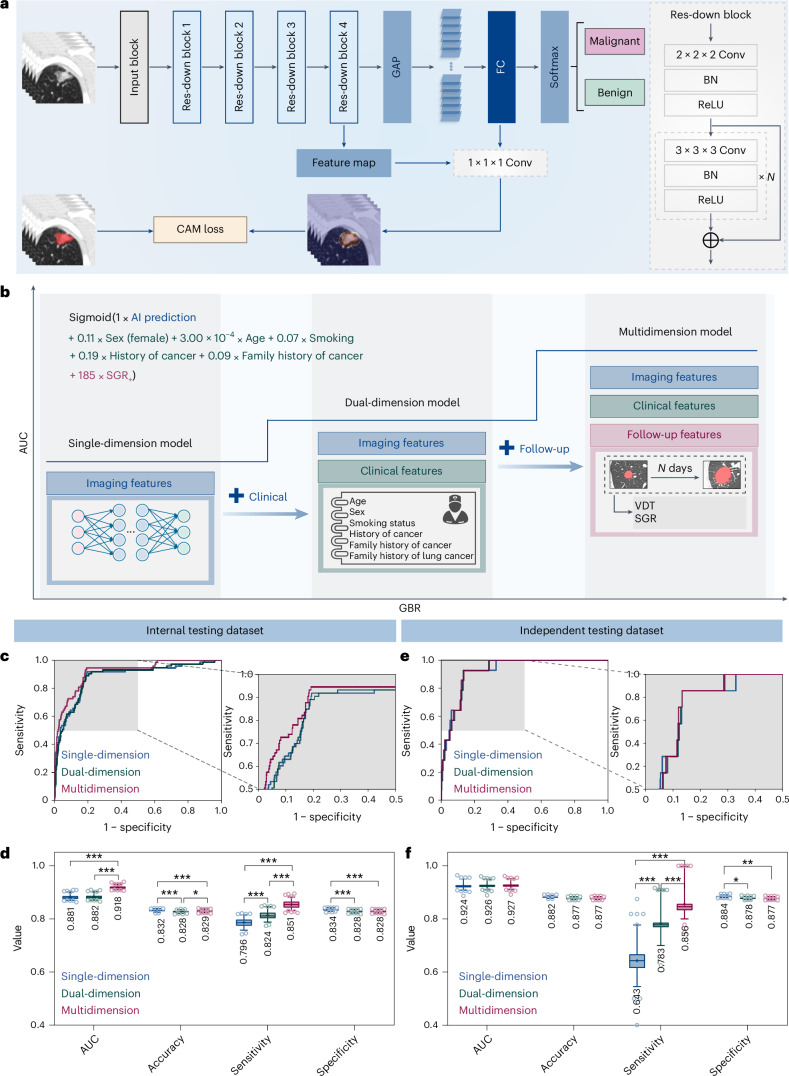
Fig. 4. GBR model incorporating multidimensional information in phase 2 used to identify the malignant nodules.
a, A DCNN was used to differentiate malignant nodules from the benign ones. b, The AUC of the GBR model increased with algorithm iterations. The GBR model based solely on AI-predicted malignant probability was refined with clinical information and follow-up features to achieve more precise risk levels. c–f, The discriminatory performance of single-, dual- and multidimensional GBR models in the internal testing dataset (c and d) and independent testing dataset (e and f) was visualized. ROC curves of the three models (c and e), with specified regions zoomed in. Quantitative metrics (d and f) of the three models in classifying malignant nodules. All metrics are shown in box and whisker plots, in which the line and the plus sign represent the median and the mean values, respectively. The numbers in the plot are the mean values. The whiskers range from the 2.5th to the 97.5th percentile, and points below and above the whiskers are drawn as individual dots. Statistical analyses in d and f were performed using ordinary two-way ANOVA followed by Tukey’s multiple comparison tests, with n = 100 replicates per condition. The asterisks represent two-tailed adjusted P values, with *P < 0.05, **P < 0.01 and ***P < 0.001. The P values in d for single-dimension versus dual-dimension, single-dimension versus multidimension and dual-dimension versus multidimension were 0.451, <0.001 and <0.001 for AUC; <0.001, <0.001 and 0.025 for accuracy; <0.001, <0.001 and <0.001 for sensitivity; and <0.001, <0.001 and 1.000 for specificity, respectively. The P values in f for single-dimension versus dual-dimension, single-dimension versus multidimension and dual-dimension versus multidimension were 0.876, 0.565 and 0.857 for AUC; 0.067, 0.068 and 1.000 for accuracy; <0.001, <0.001 and <0.001 for sensitivity; and 0.012, 0.004 and 0.946 for specificity, respectively. Res, residual; Conv, convolution; FC, fully connected; BN, batch normalization; ReLU, rectified linear unit.