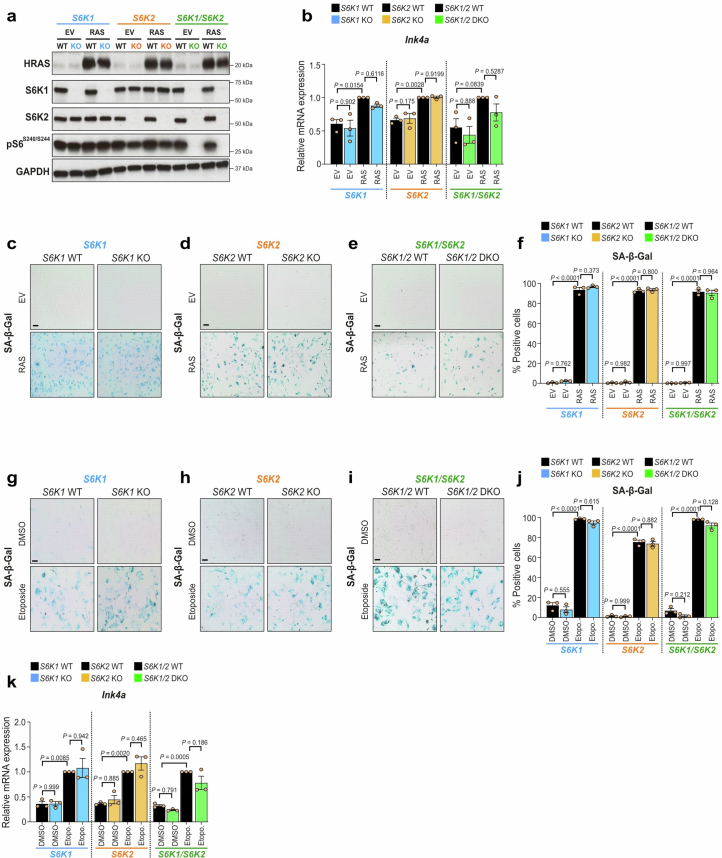
Extended Data Fig. 5. Induction of senescence in MEFs with different S6K1 and S6K2 status.
a. Immunoblot images of a single experiment for HRAS, S6K1, S6K2, pS6S240/S244 and GAPDH expression. S6K1 and GAPDH (loading control) were run on the same blot. S6K2, pS6S240/S244 and HRAS were run on separate blots; therefore, GAPDH served as a sample preparation control for those blots. b. Relative mRNA expression for Ink4a assessed by RT-qPCR from MEFs transduced with empty vector (EV) or HRASG12V (RAS) from S6K1 WT (n = 3) and KO (n = 3), S6K2 WT (n = 3) and KO (n = 3) as well as S6K1/2 WT (n = 3) and DKO (n = 3) cells. c-f. Quantification (f) and representative images (c-e) of senescence-associated beta-galactosidase (SA-β-Gal) staining in S6K1, S6K2, and S6K1/2 MEFs with the indicated genotype expressing either EV or RAS vector to undergo OIS (n = 3). g-j. Quantification (j) and representative images (g-i) of senescence-associated beta-galactosidase (SA-β-Gal) staining in S6K1, S6K2, and S6K1/2 MEFs with the indicated genotype treated with DMSO or etoposide (5 μM) to undergo TIS (n = 3). k. Relative mRNA expression for Ink4a was assessed by RT-qPCR from MEFs treated with DMSO or etoposide (5 μM) to undergo TIS (n = 3). mRNA expression was normalized to the Rps14 housekeeping gene. Data are expressed as mean ± SEM. Statistical significance was calculated using two-way analysis of variance with Tukey’s multiple comparison test. n denotes individual MEFs generated from independent embryos. WT, wild-type. KO, knockout. DKO, double knockout. Scale bar, 100 μm.