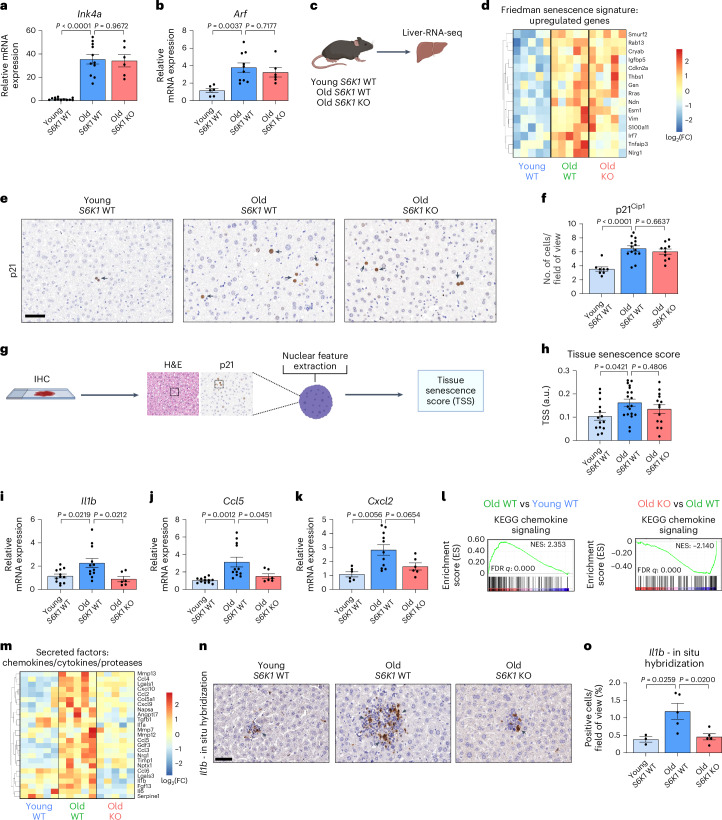
Fig. 2. S6K1 status affects inflammation but not senescence in the livers of old mice.
a,b, Relative mRNA expression for Ink4a (a) and Arf (b) were assessed by RT–qPCR from whole liver lysates of young S6K1 WT (90 d; n = 12 Ink4a and n = 6 Arf), old S6K1 WT (600 d; n = 11 Ink4a and n = 10 Arf) and old S6K1 KO (600 d; n = 6 for Ink4a and Arf) mice. mRNA expression was normalized to the Rps14 housekeeping gene. c, Experimental scheme. Bulk RNA-seq of whole liver lysates from young S6K1 WT (90 d), old S6K1 WT (600 d) and old S6K1 KO (600 d) mice. d, Heatmap depicting the expression of key regulated genes in the ‘Friedman senescence signature’ in young S6K1 WT (90 d; n = 5), old S6K1 WT (600 d; n = 5) and old S6K1 KO (600 d; n = 5) mice. e,f, p21 staining (e) and quantification (f) of young S6K1 WT (90 d; n = 8), old S6K1 WT (600 d; n = 14) and old S6K1 KO (600 d; n = 10) mice. Scale bar, 50 µm. g, Pipeline for calculating TSSs based on nuclear parameter extraction from H&E-stained liver tissue slides. h, TSSs of young S6K1 WT (90 d, n = 14), old S6K1 WT (600 d, n = 19) and old S6K1 KO (600 d, n = 14) mice. i–k, Relative mRNA expression for Il1b (i), Ccl5 (j) and Cxcl2 (k) assessed by RT–qPCR from whole liver lysates of young S6K1 WT (90 d; n = 12 for Il1b and Ccl5; n = 6 for Cxcl2), old S6K1 WT (60 d; n = 12 for Il1b and Ccl5; n = 11 for Cxcl2) and old S6K1 KO (60 d; n = 6 for Il1b, Ccl5 and Cxcl2) mice. mRNA expression was normalized to the Rps14 housekeeping gene. l, GSEA for ‘KEGG Chemokine Signaling’ of young S6K1 WT (90 d), old S6K1 WT (600 d) and old S6K1 KO (600 d) mice from whole liver lysates. m, Heatmap depicting the expression of key chemokines, cytokines and proteases in young S6K1 WT (90 d; n = 5), old S6K1 WT (600 d; n = 5) and old S6K1 KO (600 d; n = 5) mice. n,o, In situ hybridization for Il1b mRNA (n) and quantification (o) in young S6K1 WT (90 d; n = 3), old S6K1 WT (600 d; n = 5) and old S6K1 KO (600 d; n = 5) mice. Scale bar, 50 µm. Data are expressed as mean ± s.e.m. Statistical significance was calculated using one-way ANOVA with Tukey’s multiple comparison test. n denotes individual mice. FC, fold change; FDR, false discovery rate; IHC, immunohistochemistry; NES, normalized enrichment score.