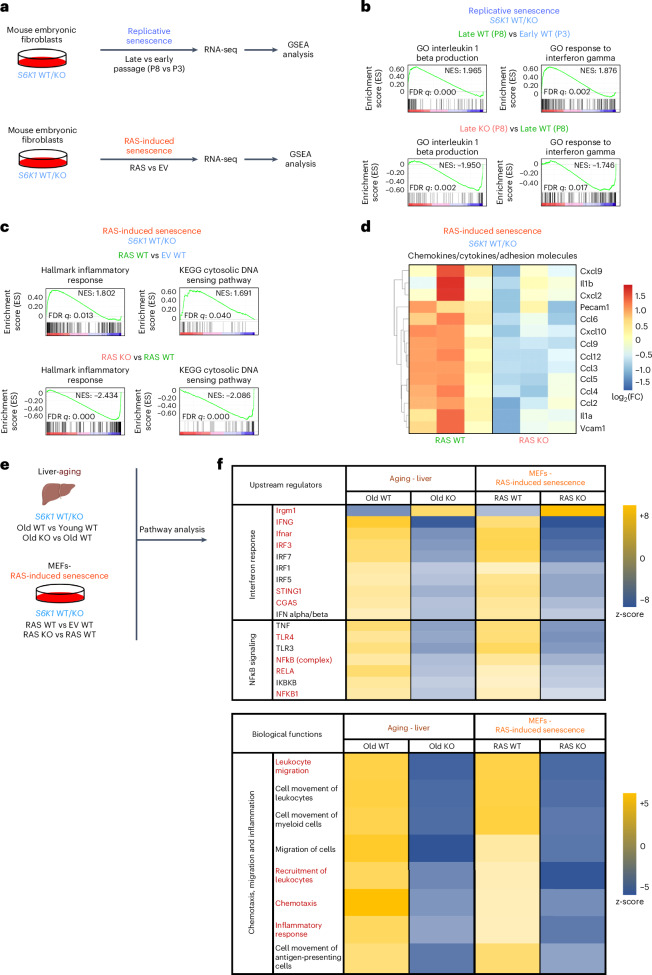
Fig. 6. Transcriptional analysis shows that S6K1 regulates inflammatory pathways.
a, Experimental scheme. MEFs from S6K1 WT/KO embryos were assessed for replicative senescence or RAS-induced senescence. Samples underwent subsequent RNA-seq and GSEA. b. GSEA of early S6K1 WT (passage 3), late S6K1 WT (passage 8) and late S6K1 KO (passage 8) MEFs. c, GSEA of S6K1 WT MEFs expressing an EV, S6K1 WT MEFs expressing RASG12V or S6K1 KO MEFs expressing RASG12V. d, Heatmap illustrating the gene expression pattern of key pro-inflammatory SASP factors involved in RAS-induced senescence. Left, comparison of S6K1 WT MEFs expressing RASG12V (n = 3) with S6K1 WT MEFs expressing EV (n = 3). Right, comparison of S6K1 KO MEFs expressing RASG12V (n = 3) with S6K1 WT MEFs expressing RASG12V (n = 3). e, Schematic of combined pathway analysis of the aging cohort and in MEFs undergoing RAS-induced senescence of the indicated comparisons to identify common upstream regulators and biological functions. f, Top, assessment of common upstream regulators of the SASP in S6K1 KO mice in the aging liver and S6K1 KO MEFs undergoing RAS-induced senescence. Bottom, assessment of biological functions that are commonly regulated in S6K1 KO mice in the aging liver and in S6K1 KO MEFs undergoing RAS-induced senescence. FC, fold change; FDR, false discovery rate; NES, normalized enrichment score; P, passage.