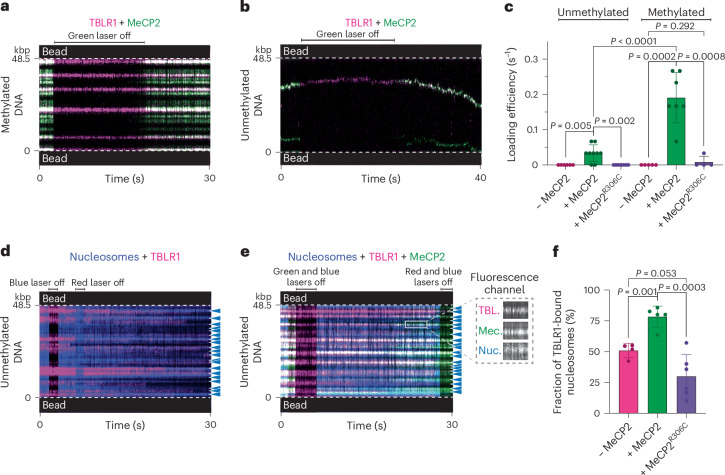
Fig. 6. MeCP2 mediates effector recruitment to methylated DNA and nucleosomes.
a,b, Representative kymographs of a CpG methylated (a) or unmethylated (b) DNA tether incubated with Cy3-labeled MeCP2 and LD655-labeled TBLR1. c, Loading efficiency for 20 nM LD655-TBLR1 binding in the absence of MeCP2 to unmethylated (n = 6 independent tethers) or methylated (n = 5 independent tethers) DNA, in the presence of 2 nM MeCP2 to unmethylated (n = 9 independent tethers) or methylated (n = 7 independent tethers) DNA, or in the presence of 2 nM MeCP2R306C to unmethylated (n = 7 independent tethers) or methylated (n = 4 independent tethers) DNA. Each dot represents data from one independent tether. The bars represent mean and s.d. d,e, Representative kymographs of an AF488-nucleosome-containing unmethylated DNA tether incubated with LD655-TBLR1 only (d) or with both LD655-TBLR1 and Cy3-MeCP2 (e). Lasers were switched on and off to confirm fluorescence signals from each channel. The arrows denote nucleosome positions. The inset shows a zoom-in view of individual fluorescence channels at a nucleosome site (Nuc.) where TBLR1 (TBL.) and MeCP2 (Mec.) colocalized. f, Fraction of nucleosomes loaded on an unmethylated DNA tether that were colocalized with TBLR1 in the absence (n = 4 independent tethers) or presence of 2 nM MeCP2 (n = 5 independent tethers) or MeCP2R306C (n = 6 independent tethers). Each dot represents data from one independent tether. The bars represent mean and s.d. Significance for c and f was calculated using a two-tailed unpaired t-test for each pair.