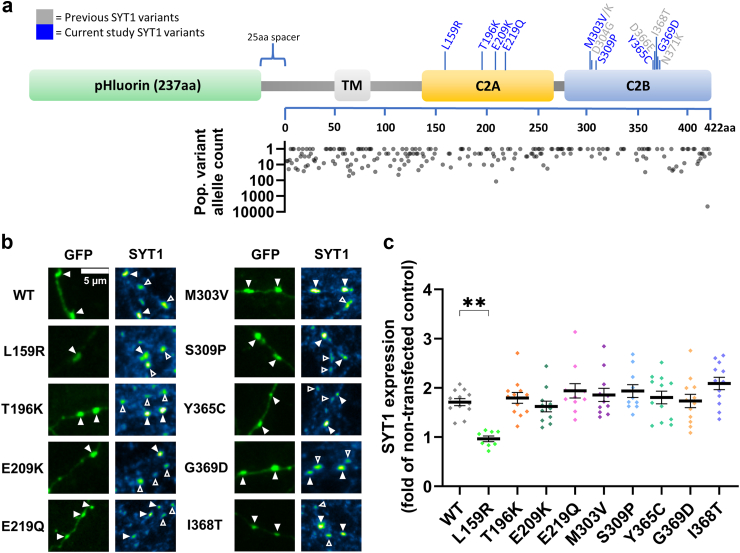
Fig. 1.
SYT1 variants, except L159R, are expressed as efficiently as the WT protein at presynaptic nerve terminals. Hippocampal neurons co-transfected with SYT1-pHluorin variants and empty vector pcDNA3.1 were fixed and immunolabelled for GFP (i.e. pHluorin, to identify transfected neurons) and SYT1. (a) Linear protein sequence of SYT1-pHluorin construct highlighting the location of previously investigated (grey) and currently examined (blue) SYT1 variants. Graph indicates allele frequency of population missense variants in SYT1 from GNOMAD v4.10 (note logarithmic scale, average population allele read number is 1,611,866). No population missense variants are present at putative pathogenic residues. (b) Representative images of immunolabelled neurons transfected with SYT1 variants (tagged with pHluorin). Note that WT refers to cells transfected with wild-type SYT1-pHluorin. Left panels show SYT1-pHluorin-transfected neurons in green and right panels display SYT1 immunofluorescence, with warmer colours indicating more intense staining. Arrowheads indicate transfected (closed) and non-transfected (open) nerve terminals. Scale bar = 5 μm. (c) SYT1 expression levels at nerve terminals, expressed as the SYT1 immunofluorescence intensity in transfected neurons, relative to non-transfected neurons in the same field of view. Data displayed as mean ± SEM, n = 8–12 (and see Supplementary Table S2a). Kruskal–Wallis test with Dunn's multiple comparison test compared to WT (n = 12); L159R p = 0.0076 (indicated by ∗∗; n = 8), T196K p > 0.99 (12), E209K p > 0.99 (11), E219Q p > 0.99 (11), M303V p > 0.99 (11), S309P p > 0.99 (10), Y365C p > 0.99 (12), G369D p > 0.99 (12), I368T p = 0.60 (12). For all experiments, ‘n’ refers to an individual field of view from an independent coverslip. All experiments were repeated across at least 3 independent cultures, with each culture comprising at least 3 embryos.