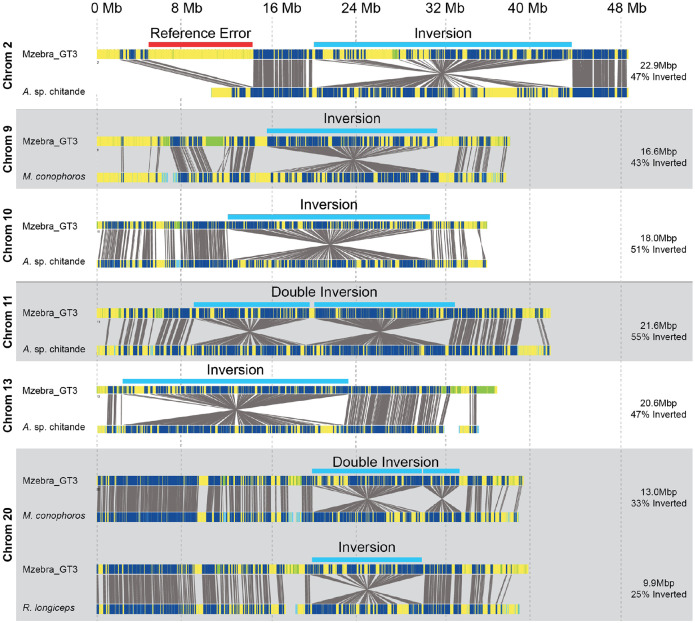
Figure 2. Identification of six large single or double inversions segregating within Lake Malawi cichlids.
Representative alignments of inversions or double inversions identified from blood samples obtained from 30 individuals from eleven species. For each inversion, the top shows predicted motifs from an in-situ digest of the reference genome, the bottom shows motifs identified using Bionano molecules obtained from an individual of the species indicated on the left, and the grey lines indicate matching motifs based upon predicted and observed distances. Single inversions were identified on chromosomes 2, 9, 10, 13, and 20. Tandem double inversions were identified on chromosomes 11 and 20. The estimated length and percentage of the chromosome spanned by the inversion is shown on the right. The single inversion on 20 identified in Rhamphochromis longiceps has the same position as the first inversion of the double inversion on 20 identified from Mchenga conophoros. A list of all samples and their inversion genotype is in Table 1. Note that an error in the reference genome in chromosome 2 is indicated in the first panel.