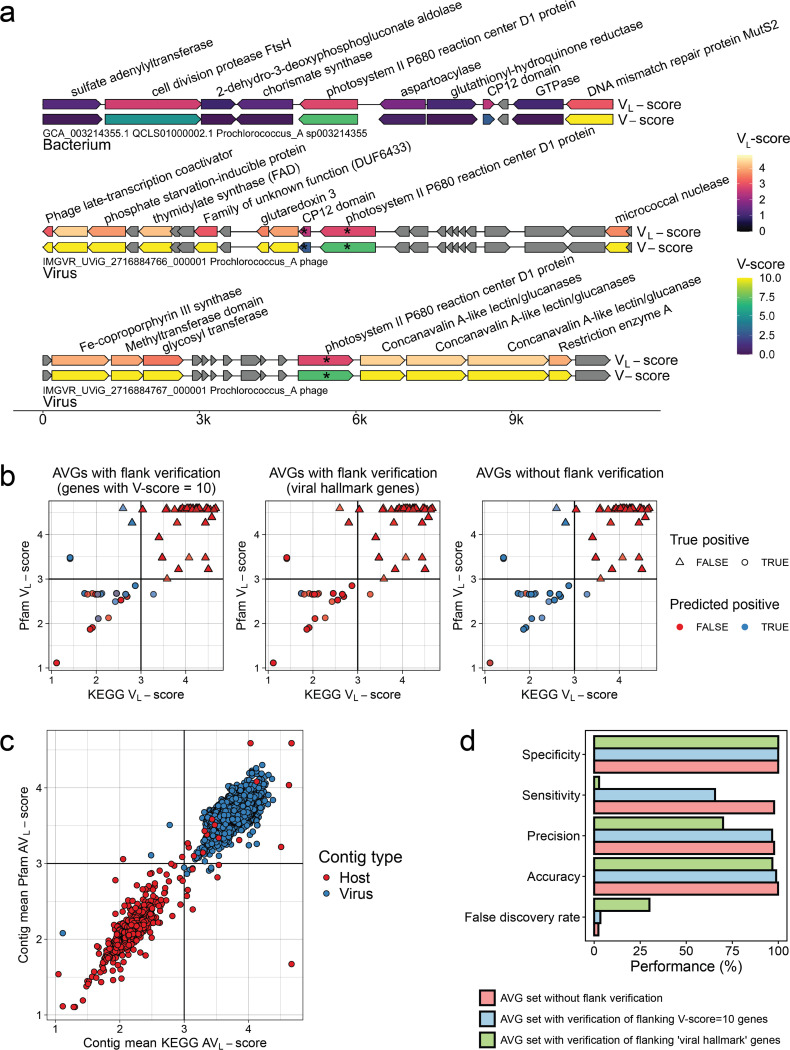
Fig. 4 |. Application of V-scores, VL-scores, and AVL-scores to auxiliary gene (AVG) detection.
a, V-scores and VL-scores reveal AMGs in viral genomes and distinguish AMGs from host-encoded metabolic genes. Genes with an asterisk (*) were predicted as AMGs using the described workflow (see Methods). b, Establishing optimal Pfam VL-score / KEGG VL-score combinations to distinguish viral auxiliary vs. non-auxiliary genes. Points represent individual genes in our database of viral and host genomes that had both Pfam5 and KEGG6 annotations matching to either the database of the 17 AMGs or 10 non-AMG protein families. Genes marked as potentially auxiliary have a maximum KEGG and Pfam VL-scores of 3, as indicated by the vertical and horizontal lines. c, Establishing the optimal Pfam/KEGG AVL-score of query scaffolds to distinguish viral vs. host genomes. Points represent individual genes, plotted by the AVL-score of all Pfam or KEGG annotations encoded by the gene’s origin scaffold. Vertical and horizontal lines represent the chosen scaffold AVL-score used to distinguish viral from host scaffolds (> 3: virus, < 3: host). Points are colored by the actual scaffold type of the gene’s origin (host or virus). d, Performance of the proposed AMG identification workflow. Performance was evaluated based on the confusion matrices in Supplementary Table S12.