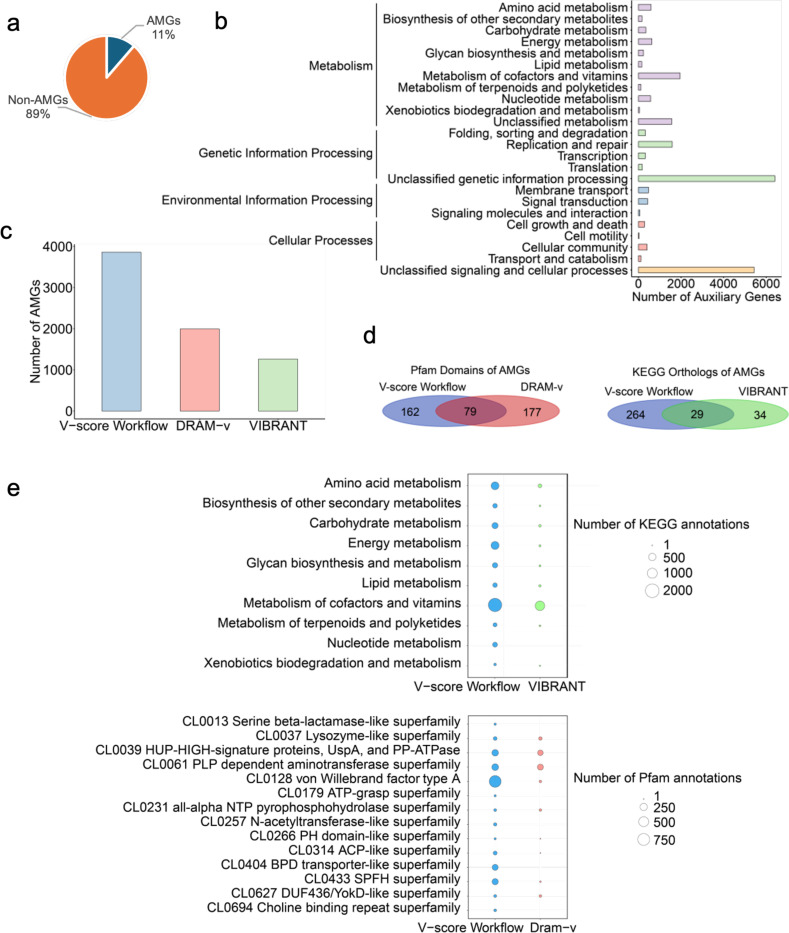
Fig. 5 |. Auxiliary genes identified in the study and comparison with existing methods.
a, auxiliary gene composition. AMG: auxiliary metabolic genes. b, Potential functions of auxiliary genes with annotations detected using the V-score workflow. Purple bars represent categories within Metabolism, green bars denote Genetic Information Processing, blue bars indicate Environmental Information Processing, pink bars correspond to Cellular Processes, and orange bars represent unclassified signaling and cellular processes. c, Number of AMGs identified by the V-score workflow compared to other existing methods, including DRAM-v and VIBRANT. d, Overlap and unique Pfam domains or KEGG orthologs of AMGs identified by the V-score workflow, DRAM-v, and VIBRANT. e, Comparison of the number of KEGG or Pfam annotations of AMGs identified using the V-score workflow, DRAM-v, and VIBRANT. Please note that VIBRANT exclusively outputs results that contain KEGG annotations, while DRAM-v mainly generated Pfam annotations for AMGs identified in the study.