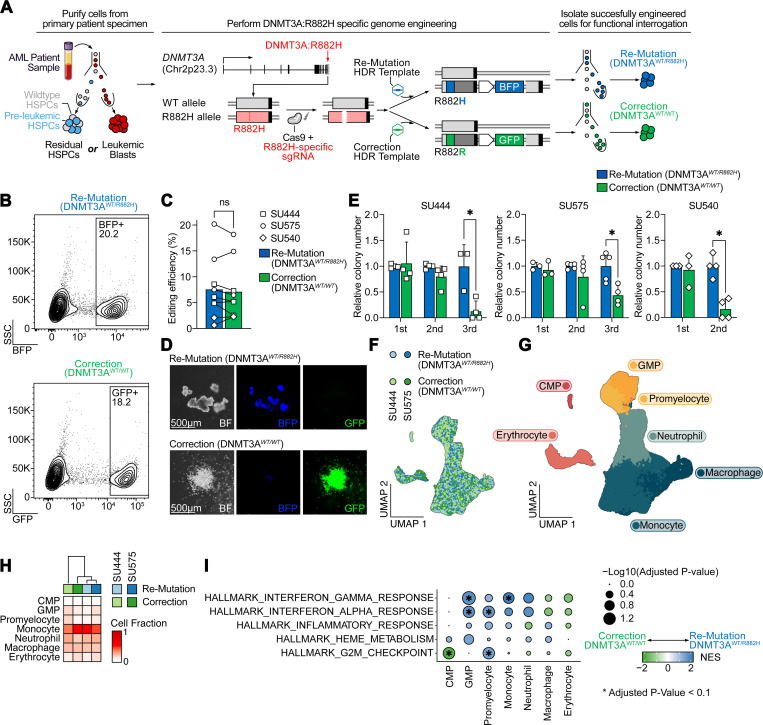
Fig. 1: Correction of DNMT3AR882H in pre-leukemic HSCs results in normalization of aberrant self-renewal.
A: Schematic of DNMT3AR882H correction approach. Cell populations of interest (either residual HSPCs containing pre-leukemic HSPCs or leukemic blasts) are purified by flow cytometry. Subsequently, pre-complexed Cas9 and sgRNA targeting the mutant, R882H allele, are electroporated into the target cells and cells are transduced with rAAV6 containing either the codon-optimized mutant (R882H) or wildtype (R882R) sequence, and a selectable fluorescent marker (BFP or GFP). Successfully engineered cells are then isolated for functional experiments. B: Isolation by FACS of successfully edited residual HSPCs (from patient SU575) for either re-mutation (top panel) or correction (bottom panel) of DNMT3AR882H. C: Editing efficiency across all experiments in residual HSPCs as percentage of fluorescent protein positive cells (n=3 independent patient specimens). D: Representative hematopoietic colonies 14 days after plating into semisolid media (Methocult). E: Serial replating assay of gene-edited residual HSPCs derived from three independent patient specimens (n=3–4 technical replicates per patient), * P < 0.05. F: UMAP embedding of single-cell RNA-Seq from gene-edited residual HSPCs differentiated in vitro for 14 days after editing colored by genotype and donor. G: UMAP embedding as in F colored by assigned cell type. H: Relative abundance of cell types within each sample. Sample identities and DNMT3A genotypes are indicated at the top. I: Gene set enrichment analysis of Hallmark gene sets between re-mutation (DNMT3AWT/R882H, blue) and correction (DNMT3AWT/WT, green) across cell types. * P < 0.1.