Figure 7. Contingency relationships between distinct peptide positions.
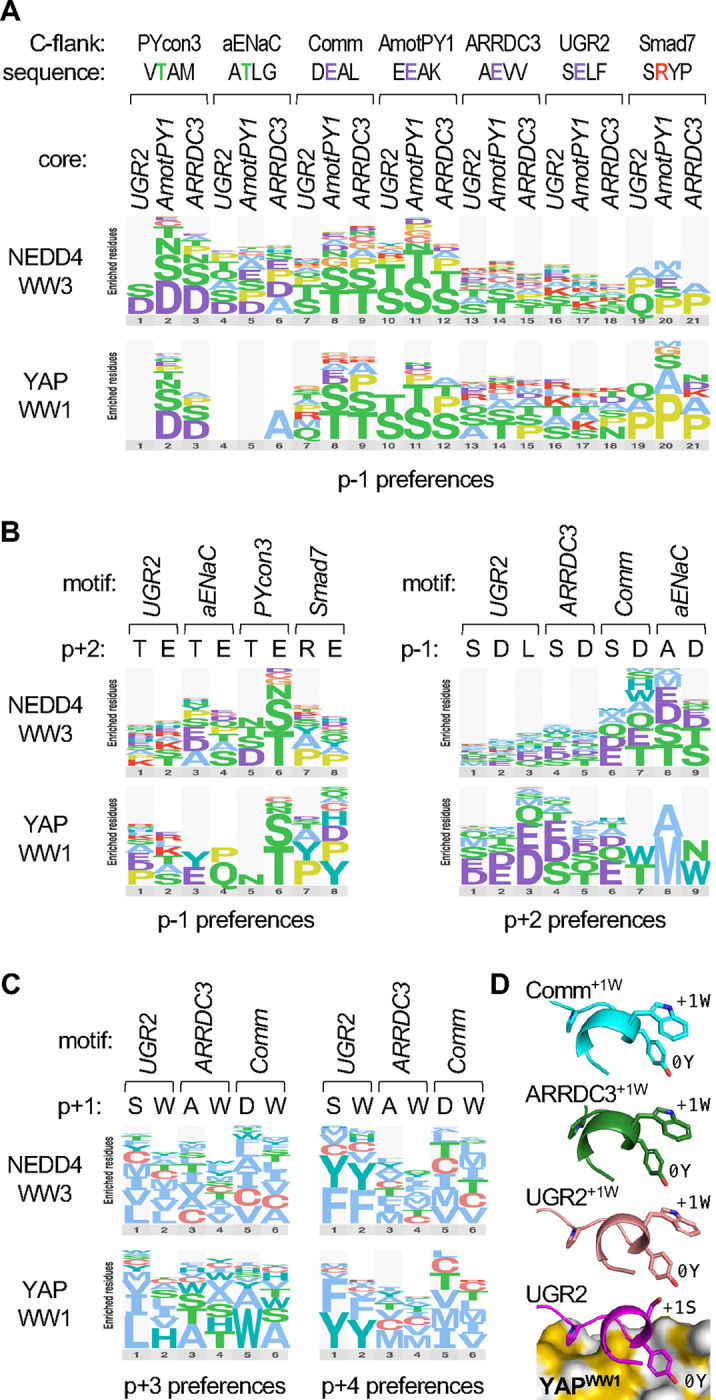
(A) Logos of p−1 preferences in 21 different peptide contexts. Seven 4-residue C-flank sequences were appended to each of three core motifs (…[LP]PxY), and then the p−1 codons were randomized and tested for binding YAPWW1 and NEDD4WW3. For clarity, the logos show only the most preferred residues, and they omit data from peptides for which the difference in z-score between the tested WW-Cln2 fusion and unfused Cln2 was less than 1.
(B) Logos showing preferences at p−1 when the p+2 residue is varied (left), or preferences at p+2 when the p−1 residue is varied (right).
(C) Logos showing preferences at p+3 or p+4 when the p+1 residue is varied. Data in panels A-C are derived from the median synonym z-scores from 3 independent experiments.
(D) Predicted conformations of peptides bound to YAPWW1. Bottom, prediction for the UGR2 peptide, showing 3 side chains for reference (−2P, 0Y, +1S). Above, analogous predictions for peptides with p+1 Trp substitutions (+1W), aligned with the bottom structure and hiding the YAPWW1 domain for clarity. The Trp groups are predicted to lie next to the p0 Tyr groups (0Y) with their planar faces in a perpendicular geometry.
