Figure 8. Stratification of [LP]PxY matches by PSSM scoring.
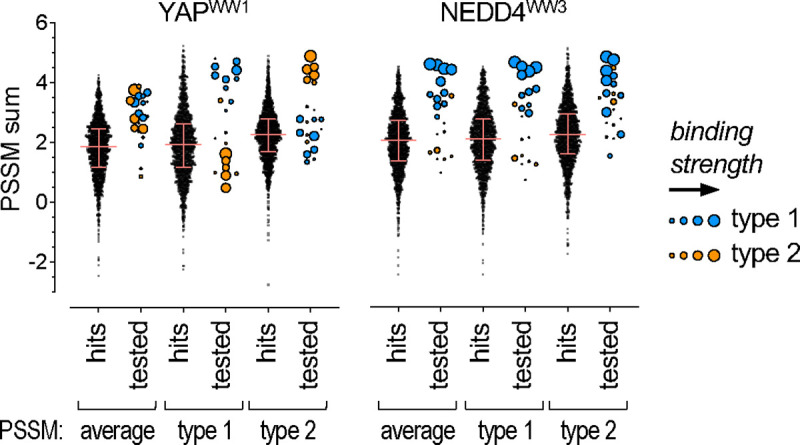
All 1730 matches to the [LP]PxY consensus sequence (“hits”) in disordered regions of the human proteome were scored using 6 different PSSMs. For each of 2 WW domains, 3 PSSMs were derived from the DMS results: one from all motifs (average), one from type 1 motifs only, and one from type 2 motifs only. The PSSM values for individual residues were summed across 10 motif positions (xx[LP]PxYxxxx) to calculate a predicted score (PSSM sum) for all sequences. For all hits, the distribution of sums obtained using each PSSM is shown; pink lines denote median and quartile values. For all motifs tested by SIMBA (“tested”), PSSM sums were calculated similarly, and their symbol sizes are proportional to their observed SIMBA scores (see Figure 6B).
