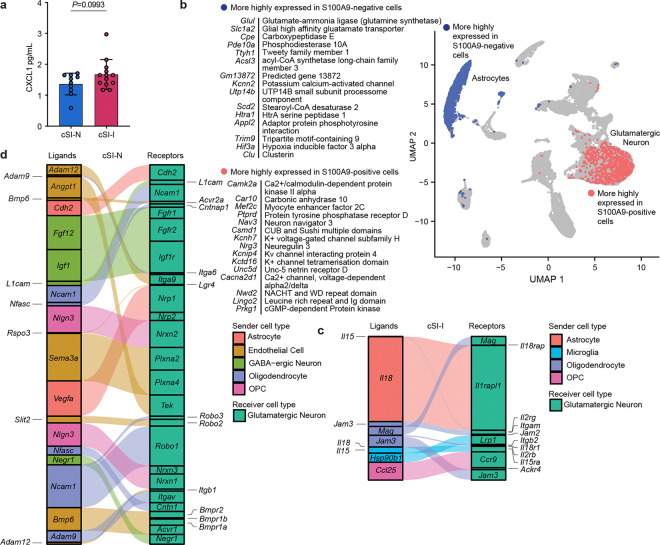
Extended Data Fig. 6. snRNA-Seq intercellular signaling and cellular response to inflammation in the cerebral cortex of P37 mice.
a, Levels of the protein CXCL1 in the cerebral cortex tissue of P37 cSI-I and cSI-N animals (n=6–13 mice/group, bars denote mean ± s.d., Welch’s t test). b, Scissor-defined relationship between serum S100A9 and snRNA-Seq gene expression. A correlation was first performed between bulk RNA-Seq gene expression and serum S100A9 levels in the same animals (n=49 P37 mice; n=15–18 mice/group); subsequently, a Gaussian regression was performed using this correlation matrix with snRNA-Seq expression (n=4 mice/group). Shown are the cortex nuclei that were positively (pink) and negatively (blue) associated with levels of systemic S100A9. Top inset: 15 genes most significantly up-regulated in astrocyte nuclei that were negatively correlated with S100A9 compared to non-associated (‘background’) astrocytes. Bottom inset: Genes most significantly differentially expressed in glutamatergic neurons that were positively correlated with S100A9 compared to non-associated (‘background’) glutamatergic neurons. c,d, Predicted NicheNet receptors (right) in glutamatergic neurons for ligands (left) expressed by various cell types in the cerebral cortex of P37 animals. c, Ligands more highly expressed in cSI-I animals; d, ligands more highly expressed in cSI-N animals. Boxes are sized proportionally to the weighted ligand/receptor score; corresponds to Fig. 3f.