Abstract
The first outbreaks of coronavirus CoV, SARS-CoV and MERS-CoV have occurred in China and Saudi Arabia over the past decade, respectively. From the end of 2019, a great battle began by the world scientific community against SARS-CoV-2, the virus that caused COVID-19, a pathology that generated devastating consequences on all existing continents. Several mutations have already been detected in the structure of the virus, which have been responsible for the generation of many types of variants since the detection of the first COVID-19 virus identified in China. The worrisome mutations arising from the first genome of SARS-CoV-2 have been intensively studied. Some mutations increase the transmissibility of the disease through Spike, the protein responsible for binding the virus in the human cell. Among the numerous strains, the most discussed are called by the WHO as “variants of concern”. This study aims to review if COVID-19 severity may be variant dependent. Our study found tree publications that associate severity of COVI-19 symptoms to different SARS-CoV-2 variants. The most part of publications do not establish which variant is being expressed during studies. More studies with this focus are needed for a better understanding of the disease and respective variants.
Keywords: Covid-19, Variant, SARS-CoV2, Severity
Background
Considering the COVID-19 pandemic that emerged in 2019, several mutations have been responsible for the generation of many types of variants of coronavirus. In this scenario, this review aims to find in the literature an association between the severity of symptoms with the different SARS-CoV-2 variants in human adults. The results found in three studies showed that the Omicron variant causes less severe disease when compared with the Delta variant, and that the Beta variant showed low hospitalization-ICU risk and hospitalization-fatality risk. However, no study presented a specific relation between the symptoms and the SARS-CoV-2 variant, just the level of the disease severity caused by it. Therefore, more studies with this focus are needed for a better understanding of the disease and respective variants.
Introduction
In 1968, under an electron microscope, a team of virologists [1] visualized a new group of viruses with a morphological feature reminiscent of the solar corona. Previously, they were classified as belonging to the myxovirus group, but it was from this more detailed study of their morphology that they were differentiated and established as a separate genus known as coronaviruses [2].
The Coronaviridae were classified as a monogenic family of ether-labile, pleomorphic enveloped viruses belonging to the order Nidovirales. This order also includes the families Arteriviridae, Mesoniviridae and Roniviridae [3, 4].
The first human coronaviruses (HCoVs) have been known since the late 1960s, when HCoV-229E and HCoV-OC43 were first isolated. These HCoVs are alpha-coronaviruses and are distributed worldwide. They can generate common cold symptoms such as headache, sneezing, malaise and sore throat that last for about two weeks and are predominantly transmitted during winter.
Nidoviral viruses have large genomes, and some have the largest recognized ribonucleic acid (RNA). Within the Nidovirus families, the main distinctions are the number, type, and size of structural proteins. Coronaviridae has about 15,000–30,000 nucleotides with a molecular weight between 5 × 106 and 7 × 106, containing a single-stranded genomic RNA with a diameter ranging from 60 to 220 nm [3, 4]. There are four subgroups in the coronavirus family: alpha, beta, gamma, and delta-coronavirus. Alpha and beta primarily originate from bats, while published studies report that gamma and delta viruses originate from pigs and birds [4].
The first coronavirus outbreak occurred in 2002 in Guangzhou, China. Studies indicate that SARS-CoV was a virus that mainly hit bats that transmitted it to a mammal called civet (the intermediate host) and this last used to infect humans. None of these species would coexist in the same place and time, however, animal markets offered this opportunity as animals were placed in stacked cages while still alive, causing fluids and droppings to leak between species [5].
Consequently, the virus was able to jump from one species to another, evolving and adapting along the way [5]. The final product was an outbreak that caused the infection of 8,096 people in 30 countries and 774 deaths with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) (about 9% of the fatality rate) [6]. It is estimated that each infected person could transmit the virus to two secondary cases. The incubation period of the disease caused by SARS-CoV can occur from 4 to 7 days and the peak of viral load occurs on the 10th day of illness [7].
In 2004, HCoV-NL63 was discovered in the Netherlands, causing symptoms such as runny nose, conjunctivitis, fever, and bronchiolitis, with a peak in early summer. Also, in 2004, HCoV-HKU1 was isolated in Hong Kong. This virus caused acute asthmatic exacerbation and mild respiratory illness [7].
Considering the mild sintomatology caused by HcoVs in the meantime, they are classified as community-acquired viruses well adapted to humans. Moreover, they are less likely to mutate and cause serious disease, are less virulent and therefore cannot easily generate a pandemic frame.
In 2012, a new human coronavirus emerged in the Middle East, generating the Middle East Respiratory Syndrome-CoV (MERS-CoV) and until August 2014 it was responsible for 855 cases with a fatality rate of almost 40%, according to the European Center for Diseases. Prevention and control [8].
Lately, SARS-Cov-2 dated December 2019, in Wuhan, China, had been identified [7].
Clinical studies showed in SARS-CoV-2 patients’ fever, shortness of breath, non-productive cough, headache, dyspnea, myalgia, fatigue, lymphopenia, radiographic evidence of pneumonia and acute lung injury. In most patients, chest CT revealed a distribution of ground-glass opacities in both lungs. This acute lung injury could often lead to hypoxemic respiratory failure, ARDS, and ultimately death [5]. Compared to the initial SARS-CoV, the transmission rate of SARS-CoV-2 is higher, and this is a result attributed to genetic recombination in S protein, in the region of the spike protein binding site of SARS-CoV-2 [9]. In late 2020, the World Health Organization prompted some criteria to variants classification, separating the variants of interest and variants of concern in order to monitor the SARS-CoV2 behaviour. In 2024 The World Health Organization (WHO) lauched the Coronavirus Network (CoViNet) to help detection and tracking the variants. In this context, the virus spread and chronically infected individuals symptoms could be observed.In march 2023 WHO uptadet this system. According to updated data from March 2024, COVID-19 caused 7,037,007 deaths globally.
In order to be considered a variant of interest, the variant must meet the following criteria working definition (updated 15 March 2023) by WHO: a SARS-CoV-2 variant with genetic changes that are predicted or known to affect virus characteristics such as transmissibility, virulence, antibody evasion, susceptibility to therapeutics and detectability; and identified to have a growth advantage over other circulating variants in more than one WHO region with increasing relative prevalence alongside increasing number of cases over time, or other apparent epidemiological impacts to suggest an emerging risk to global public health. In addition, to consider the classification “variant of concern” there is more one criterion to consider as detrimental change in clinical disease severity; or change in COVID-19 epidemiology causing substantial impact on the ability of health systems to provide care to patients with COVID-19 or other illnesses and therefore requiring major public health interventions; or significant decrease in the effectiveness of available vaccines in protecting against severe disease [10].
In the scientific field, systematic reviews are highly valued, considering that their evidence will be the focus of numerous guidelines and medical decisions. Often, however, for a reliable study, with rigorous methodological requirements, an average time interval of two years is necessary [11]. Thus, there is a need for conformity of the studies to be analyzed, to meet the answers questioned by the scientific community.
Considering the COVID pandemic that emerged in 2019, it took a global task force to provide clinical evidence with substantiated results. In this set of events, several scientific studies emerged with exceptional speed, both in terms of clinical signs, treatment, and vaccination [12].
Methodology
Currently, it is known that quick assessments are essential; not being possible to fulfill all the criteria previously established by the research groups. In this way, for example, COVID-19 studies became free in publications for viewing, reading and debates, even before peer review. This new strategy was defined as an efficient tool for the brief synthesis of new knowledge. Rapid Reviews (RRs) have been described as a type of knowledge synthesis with simplified and accelerated methods to complete the review more rapidly [13].
In 2015, the Rapid Review Methods Group (RRMG) attached to Cochrane was established and involved in research methods, including the development of standards for reporting RRs. In 2018, Cochrane’s Content Strategy identified the need to explore and, if necessary, produce RRs. To inform this work, RRMG conducted several research activities that culminated in the development of interim recommendations for RR methods. Consequently, some guidelines have been published for the elaboration of Rapid Reviews for analysis of urgent topics of higher priority [13]. The Research question of our study is:
Is it possible to associate severity of symptoms with the different SARS-CoV-2 variants in human adults?
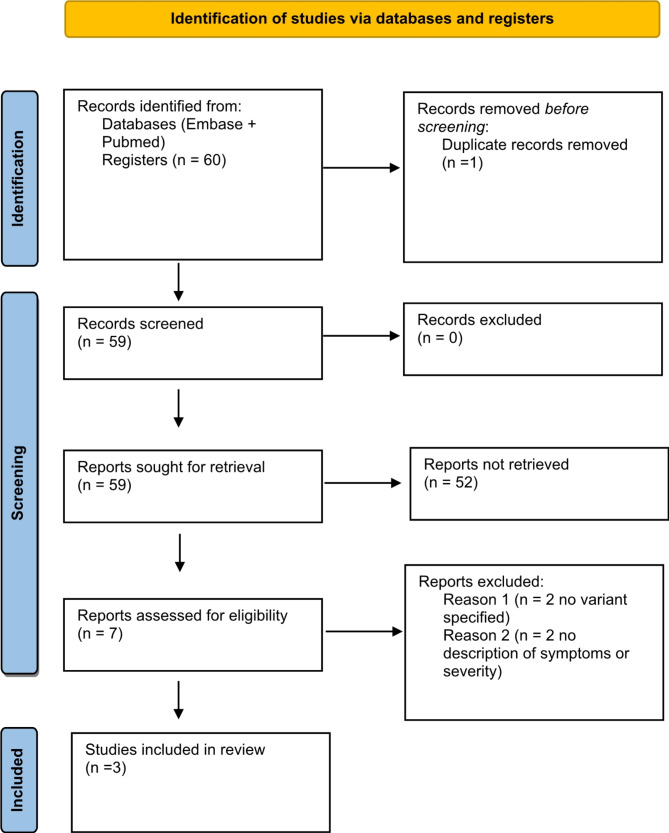
Health professionals with experience in the diagnosis and treatment of COVID-19 defined the eligibility criteria and issues of interest, in order to ensure that the research question is suitable for the interest of the scientific community. A protocol was developed with the review questions and inclusion/exclusion criteria and a diagram was developed to identify data and records, according to the suggested guideline report items for systematic reviews and meta-analyses (PRISMA), as shown in Fig. 1 [14].
Fig. 1.
PRISMA protocol
Review articles that discuss signs and symptoms of COVID-19, in adult subjects with a positive test for COVID-19, were included, looking for evaluate the severity among variants. Publications reviewed were limited to the English language, according to the suggested guidelines of the Rapid Review design instructions characteristics. Only Systematic Reviews and Meta-analyses were considered for the review, also according to the design suggestions for Rapid Review [13]. Two databases were used for research: Pubmed-MEDLINE and Embase.
PubMed-MEDLINE database
The term covid-19 as a free term presented 202,239 results. The terms “Covid-19” were used together with their synonyms rescued by Mesh terms and the term “variant” OR “variants”.
The search strategy used was:
(“COVID-19“[Mesh] OR COVID 19 OR COVID-19 Virus Disease OR COVID 19 Virus Disease OR COVID-19 Virus Diseases OR Disease, COVID-19 Virus OR Virus Disease, COVID-19 OR COVID-19 Virus Infection OR COVID 19 Virus Infection OR COVID-19 Virus Infections OR Infection, COVID-19 Virus OR Virus Infection, COVID-19 OR 2019-nCoV Infection OR 2019 nCoV Infection OR 2019-nCoV Infections OR Infection, 2019-nCoV OR Coronavirus Disease-19 OR Coronavirus Disease 19 OR 2019 Novel Coronavirus Disease OR 2019 Novel Coronavirus Infection OR 2019-nCoV Disease OR 2019 nCoV Disease OR 2019-nCoV Diseases OR Disease, 2019-nCoV OR COVID19 OR Coronavirus Disease 2019 OR Disease 2019, Coronavirus OR SARS Coronavirus 2 Infection OR SARS-CoV-2 Infection OR Infection, SARS-CoV-2 OR SARS CoV 2 Infection OR SARS-CoV-2 Infections OR COVID-19 Pandemic OR COVID 19 Pandemic OR COVID-19 Pandemics OR Pandemic, COVID-19) AND (variant OR variants).
After the strategy, the filters of 19 years or older, human, English language, Systematic Review and Meta-analysis, years: 2020–2021 were inserted. As results, 60 articles were observed.
Embase database
The free term Covid-19 presented 392,496 results. The term “COVID-19” as a free term in Embase was referred to the most used synonym, through the controlled vocabulary descriptor Emtree: “coronavirus disease 2019”.
2019 novel coronavirus disease; 2019 novel coronavirus epidemic; 2019 novel coronavirus infection; 2019-nCoV disease; 2019-nCoV infection; coronavirus disease 2; coronavirus disease 2010; coronavirus disease 2019 pneumonia; coronavirus disease-19; coronavirus infection 2019; COVID; COVID-19; COVID 19 induced pneumonia; COVID 2019; COVID-10; COVID-19; COVID-19 induced pneumonia; COVID-19 pneumonia; COVID-19; nCoV 2019 disease; nCoV 2019 infection; new coronavirus pneumonia; novel coronavirus 2019 disease; novel coronavirus 2019 infection; novel coronavirus disease 2019; novel coronavirus infected pneumonia; novel coronavirus infection 2019; novel coronavirus pneumonia; paucisymptomatic coronavirus disease 2019; SARS coronavirus 2 infection; SARS coronavirus 2 pneumonia; SARS-CoV-2 disease; SARS-CoV-2 infection; SARS-CoV-2 pneumonia; SARS-CoV2 disease; SARS-CoV2 infection; SARSCoV2 disease; SARSCoV2 infection; severe acute respiratory syndrome 2; severe acute respiratory syndrome 2 pneumonia; severe acute respiratory syndrome coronavirus 2 infection; severe acute respiratory syndrome coronavirus 2019 infection; severe acute respiratory syndrome CoV-2 infection; Wuhan coronavirus disease; Wuhan coronavirus infection.
The search strategy used was the following: (‘coronavirus disease 2019’/exp OR.
‘coronavirus disease 2019’) AND (‘variant’/exp OR variant) AND ([systematic review]/lim OR [meta analysis]/lim) AND [english]/lim AND ([adult]/lim OR [aged]/lim OR [very elderly]/lim) AND [humans]/lim AND [embase]/lim AND [2020–2021]/py. After the strategy, the filters of 18 years or older, human, English language, Systematic Review and Meta-analysis, years: 2020–2021 were inserted. Ten articles were observed as results. It was established that only studies that specified the symptom with the respective COVID-19 variant would be selected. Initially, two reviewers selected the studies for reading and analysis of inclusion and exclusion, having a third reviewer to analyze only the excluded abstracts and a fourth reviewer to analyze only the selected abstracts. Studies that could explore the symptoms generated by each variant were evaluated.
Results
A total of 60 studies was found, as shown in Fig. 1. One of these was excluded as it was a duplicate. From the remaining 59 studies, 52 were excluded after reading their title and abstract. Seven studies remained, of which 4 were excluded after reading their full text. The results found in the 3 studies included are described beneath. All included research paper answered if there are differences in severity across SARS-CoV-2 variants. Main findings are summarized in Table 1.
Table 1.
Findings summary by author
| Article (author/year) | Summary of Findings |
|---|---|
| Yu et al., 2022 | Omicron and Delta infections were found nonsevere in 97.9% and 91.4% respectively. Both infections were found to be asymptomatic in 25.5% and 8.4% of the cases respectively. |
| Wiedermann et al., 2023 |
Omicron: People up to 20 years of age mostly experienced only mild infection. Alson Omicron was found to be less severe than Delta. Alpha and Delta had similar risks in children for experiencing symptoms after 28 days of infection. Gamma variant was reported to cause more severe infections when compared to pre-VOC variants. |
| Yuan et al., 2023 |
Delta variant had the highest severity among other variants. Omicron had the lowest severity. Beta had high risk of hospitalization but low risk of hospitalization-fatality and ICU hospitalization. |
Yu et al. 2022 [15] reports that asymptomatic SARS-CoV‐2 Omicron infection is 25.5% (95% CI 17.0–38.2%), and the pooled proportion of nonsevere disease is 97.9% (95% CI). The proportion of asymptomatic Omicron infection and nonsevere disease is significantly higher than those of Delta variant 8.4% (95% CI 4.4–16.2%) and 91.4% (95% CI 87.0–96.0%) respectively. The study was conducted in five different groups, dived into: 18–65 years, pregnants, 0–20 years (children and adolescents), patient with solid organ transplant recipients and > 60 years (elderly); and another division based on the symptomatology as: asymptomatic, nonsevere COVID-19 (patients with the absence of any criteria for severe or critical COVID-19), severe COVID-19 (patients requiring oxygen saturation < 90% on room air, signs of severe respiratory distress or expressing signs of pneumonia) and critical COVID-19 (patients meeting the criteria for acute respiratory distress syndrome, having sepsis,, septic shock, or other conditions that would require the provision or life-sustaining therapies such as vasopressor or mechanical ventilation therapy).
Wiedernmann et al., 2023 [16] found that SARS-CoV-2, in people up to 20 years of age infected with Omicron mostly experienced mild disease and the symptoms experienced did not vary widely between other variants of concern (VOCs). The authors also reported that while Alpha does not appear to result in more severe COVID-19 disease, both Alpha and Delta had similar risks in children for experiencing symptoms after 28 days of infection. Compared to other variants, Gamma had more severe infections (compared with pre-VOC variants). Also, Omicron was less severe than Delta variant. The authors classified the patient’s symptomatology as: mild or asymptomatic disease (not hospitalized), moderate disease (hospitalized, but no require of intensive care|), and severe disease (hospitalized and with necessity of intensive care).
Yuan et al., 2023 [17] found in their meta-analysis the Delta variant as having the highest severity among the other variants (hospitalized, admitted to the ICU, or died). Omicron was described as having the lowest severity. Beta: had high risk of hospitalization but low risk of Intensive Care Unit (ICU) hospitalization and low hospitalization-fatality. Of all variants, Yuan et al., 2023 described the following findings: hospitalization risk: 1.51% Omicron, 4.02% Alpha, 6.56% Delta, 19.96% Beta. Fatality risk: 0.04% Omicron, 0.22% Beta, 0.46% Delta, 0.66%% Alpha. Hospitalization-fatality risk: 1.11% Beta, 3.18% Omicron, 9.06% Delta, 12.04% Alpha. Hospitalization-ICU risk: 6.01% Omicron, 15.56% Beta, 19.63% Delta, 19.99% Alpha. To measure the clinical severity of the COVI-19 disease they separated the population into case-hospitalization risk (cCHR), confirmed case-fatality risk (cCFR), hospitalization-fatality risk (HFR), and hospitalization-ICU risk (HIR).
Discussion
In this review, although none of the studies examined presented a specific relation between the symptom and the SARS-CoV-2 variant, just the level of the disease severity caused by it, was possible to conclude that the Omicron variant causes less severe disease when compared with the Delta variant, and that the Beta variant showed low hospitalization-ICU risk and hospitalization-fatality risk.
Additionally, among the studies found, Meng, et al. 2021 showed that the D614G spike variant was seen as one of the main pathogenic points related to COVID-19-induced anosmia [18]. In the same symptomatic context, Von Bartheld, et al. 2023 [19] demonstrated that Omicron variant effect on olfaction (loss of smell) is 2–10 times lower than Alpha or Delta variants.
The evidence reflects the lack of information about each strain acquired by patients and how some mutations can generate a specific symptom. Population are nowadays being unduly informed by media based on unchecked information. The most part of health services are considered misinformed about which variant is detected in COVID tests and how society should proceed with each strain, or which pathologic sign should really concern. Medical services and professionals in this field still do not have reliable means of differentiating the COVID variants in an outpatient setting, nor even obtain post-qPCR information regarding which variant is presenting prevalent among patients in the different regions of the world. The genetic mapping test to identify the mutations found in each variant of concern has a high financial impact, as it requires specialized labor and time in the results (11). In this context, it becomes almost impossible to know exactly the SARS-CoV-2 variant in each patient. Considering this scenario, unfortunately, the scientific community remains unanswered regarding the maintenance of the existence of the different strains detected of coronavirus.
Nonetheless, despite the knowledge about the variant, it is very important to evaluate age, comorbidities, and the knowledge about the patient’s vaccination status to correlate it with their clinical manifestation. Wiedernmann et al., 2023 [16] showed that mostly individuals < 18 years old infected with the omicron variant experienced mild disease, and those with ≥ 1 comorbidity was more likely to have moderate or severe disease. Furthermore, Yuan et al., 2023 [17] also described that younger people (0–24 years) demonstrated lower clinical severities when compared with the other age groups, and adults over 65 years old, possibly because they present more chronic diseases and other medical conditions, have higher clinical severity levels.
In this context of the vaccination status, Yu et al., 2022 [15] demonstrated that the proportion of asymptomatic infection and nonsevere disease in the individuals who received booster vaccines (third or the fourth dose of COVID-19 vaccination for at least 14 days) was significantly higher than the unvaccinated individuals. Thus, it is possible to observe that, besides the variant involved, all these factors: age, comorbidities and vaccination status are very important and influence directly the clinical manifestation of the SARS-CoV-2 infection. The vaccines already produced can protect from the initial variants, but even vaccinated patients are not totally immune to any of the variants [10]. It is important to notice that while this review focuses on systematic reviews and meta-analyses, future reviews/meta-analyses including observational studies and case reports may provide a broader perspective. In this scenario, it is observed the need to sum more studies to elucidate COVID-19 variants.
Conclusion
Interestingly, it was possible to observe the great scarcity of data published in systematic studies differentiating each variant, according to its symptoms.
Finally, even today there is not a conclusive discussion about the routine incorporation of the virus detection test with differentiation of the specific strain recognized. It is possible that many strains of SARS-CoV2 remain unmapped for a long time, making the decisions among quarantine and safety somehow superficial. Scientific communities should prioritize the mapping of variants of concern for the virus, so that there is a systematic and standardized precaution in relation to the real dangers that each strain can cause. More studies on the detection of each variant are of paramount importance for the improvement of approaches directed to this topic.
Acknowledgements
Not applicable.
Abbreviations
- (HCoVs)
Human coronaviruses
- (RNA)
Ribonucleic acid
- (SARS-CoV)
Severe Acute Respiratory Syndrome Coronavirus
- (MERS-CoV)
Middle East Respiratory Syndrome-CoV
- (CoViNet)
Coronavirus Network
- (WHO)
World Health Organization
- (RRs)
Rapid Reviews
- (RRMG)
Rapid Review Methods Group
- (VOCs)
Variants of concern
- (ICU)
Intensive Care Unit
- (cCHR)
Case-hospitalization risk
- (cCFR)
Confirmed case-fatality risk
- (HFR)
Hospitalization-fatality risk
- (HIR)
Hospitalization-ICU risk
Author contributions
Conceptualization, F.A.G.; mesh terms design, F.A.G., R.P., and A.A.; on-line database research and articles screening, M.D.F., M.B.B., R.P., and A.A.; article selection, F.A.C.X. and G.G.Z.; writing – original draft preparation, F.A.G., R.P., and A.A; writing – review and editing, F.A.G., M.D.F., M.B.B., F.A.C.X., D.R.M.; supervision, D.C.M.
Funding
This research received no external funding.
Data availability
Data is available from the corresponding author under reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tyrrell DA, Almeida JD, Cunningham CH, Dowdle WR, Hofstad MS, McIntosh K, et al. Coronaviridae Intervirology. 1975;5(1–2):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIntosh K. Coronaviruses: a comparative review. Current topics in microbiology and immunology / Ergebnisse Der Mikrobiologie Und Immunitätsforschung (1974): 85–129.
- 3.Siddell SG, Anderson R, Cavanagh D, Fujiwara K, Klenk HD, Macnaughton MR, et al. Coronaviridae Intervirology. 1983;20(4):181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nokhodian Z, Ranjbar MM, Nasri P, Kassaian N, Shoaei P, Vakili B, et al. Current status of COVID-19 pandemic; characteristics, diagnosis, prevention, and treatment. J Res Med Sci. 2020;25:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster RG. Wet markets–a continuing source of severe acute respiratory syndrome and influenza? Lancet. 2004;363(9404):234–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenzel RP, Bearman G, Edmond MB. Lessons from severe acute respiratory syndrome (SARS): implications for infection control. Arch Med Res. 2005;36(6):610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie M, Chen Q. Insight into 2019 novel coronavirus - an updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020;94:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erol A. Are the emerging SARS-COV-2 mutations friend or foe? Immunol Lett. 2021;230:63–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization (WHO)¹ (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). Accessed 10 December 2021.
- 11.Ganann R, Ciliska D, Thomas H. Expediting systematic reviews: methods and implications of rapid reviews. Implement Sci. 2010;5:56. [DOI] [PMC free article] [PubMed]
- 12.World Health Organization (WHO)². (https://www.who.int/news-room/feature-stories/detail/how-do-vaccines-work). Accessed 10 December 2021.
- 13.Garritty C, Gartlehner G, Nussbaumer-Streit B, King VJ, Hamel C, Kamel C, et al. Cochrane Rapid Reviews Methods Group offers evidence-informed guidance to conduct rapid reviews. J Clin Epidemiol. 2021;130:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page MJ, Moher D. Evaluations of the uptake and impact of the Preferred reporting items for systematic reviews and Meta-analyses (PRISMA) Statement and extensions: a scoping review. Syst Rev. 2017;6(1):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.YU W, GUO Y, ZHANG S, KONG Y et al. Proportion of asymptomatic infection and nonsevere disease caused by SARS-CoV-2 Omicron variant: a systematic review and analysis. J Med Virol, 94, n. 12, pp. 5790–801, Dec 2022. [DOI] [PMC free article] [PubMed]
- 16.WIEDENMANN M, IPEKCI AM, ARAUJO-CHAVERON L, PRAJAPATI N et al. SARS-CoV-2 variants of concern in children and adolescents with COVID-19: a systematic review. BMJ Open, 13, n. 10, p. e072280, Oct 9 2023. [DOI] [PMC free article] [PubMed]
- 17.YUAN Z, SHAO Z, MA L, GUO R. Clinical Severity of SARS-CoV-2 Variants during COVID-19 Vaccination: A Systematic Review and Meta-Analysis. Viruses, 15, n. 10, Sep 26 2023. [DOI] [PMC free article] [PubMed]
- 18.MENG X, PAN Y. Sep 29 COVID-19 and anosmia: the story so far. Ear Nose Throat J, p. 1455613211048998, 2021. [DOI] [PubMed]
- 19.VON BARTHELD CS, WANG L. Prevalence of olfactory dysfunction with the Omicron variant of SARS-CoV-2: a systematic review and Meta-analysis. Cells, 12, n. 3, Jan 28 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is available from the corresponding author under reasonable request.