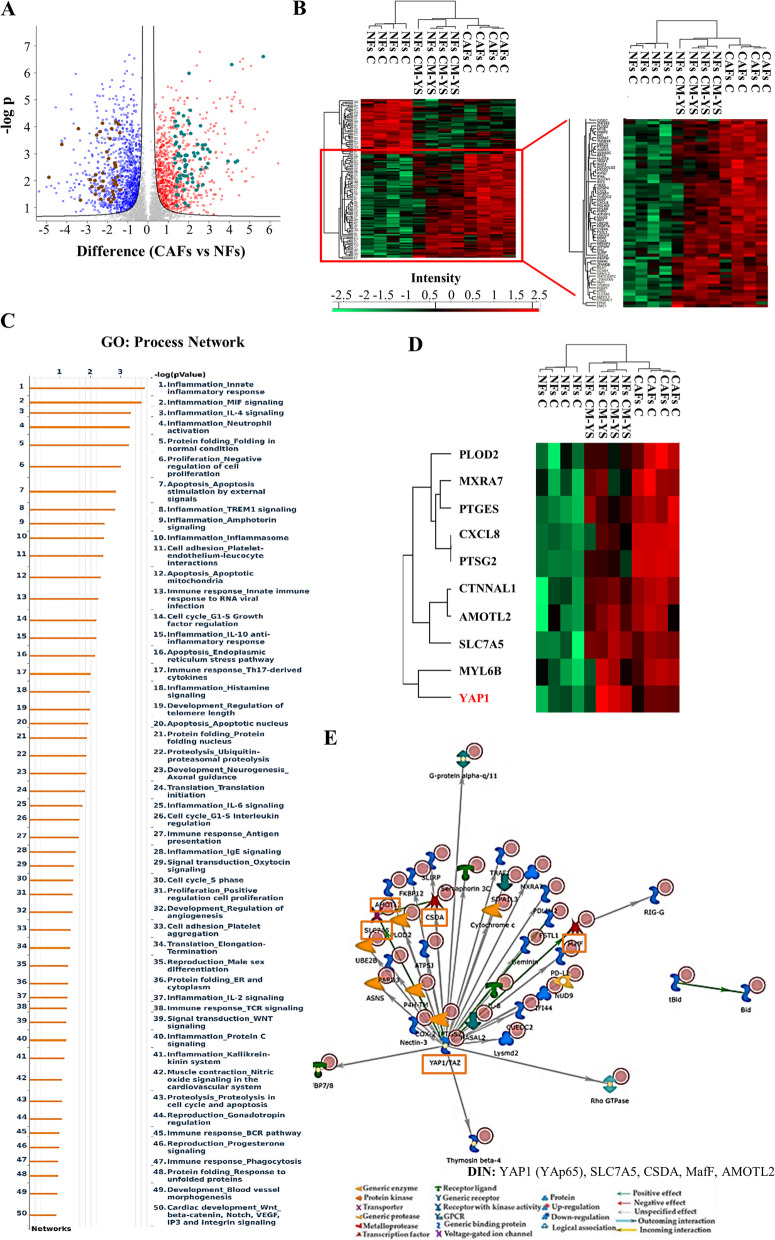
Fig. 5.
Proteomic analysis revealed proteomic similarity between NFs exposed to YS-CM and CAFs. A Volcano plot of representing all detected proteins in the comparison CAFs vs NFs. The significant proteins are localized on the right (red dots, upregulated proteins) and on the left (blue dots, downregulated proteins). Gray dots correspond to not significantly proteins, accordingly with selected parameters. This Volcano plot includes among the up (light green dots) and downregulated proteins (brown dots) those shared with NFs following their exposure to MCF-7 YS-CM reflecting in such condition their transition into CAF phenotype. B Heatmap representing supervised hierarchical clustering of the differentially expressed proteins (Fold cutoff ± 2.5) between NFs treated with CM derived from MCF-7 YS for 5 days and CAFs vs NFs identified by LC–MS/MS analysis. The heatmap is shown in red for the degree of upregulated proteins and in green for the degree of downregulated proteins (C). C GO enrichment in Process Networks of upregulated proteins (Fold cutoff ≥ 2.5) was shown. D Heatmap of the most representative upregulated proteins (Fold cutoff ≥ 2.5) of the different functional categories reported in the GO “process network”. E Direct interaction network constructed by processing the upregulated proteins identified in the comparison between NFs treated with CM derived from MCF-7 YS vs NFs (Fold cutoff ≥ 2.5). The colors of the arrows indicate positive (green), negative (red) or unknown (gray) effects of the different network items represented with different symbols (reported in the legend). DIN (orange boxes): YAP1 (YAp65), SLC7A5, CSDA, MafF, AMOTL2