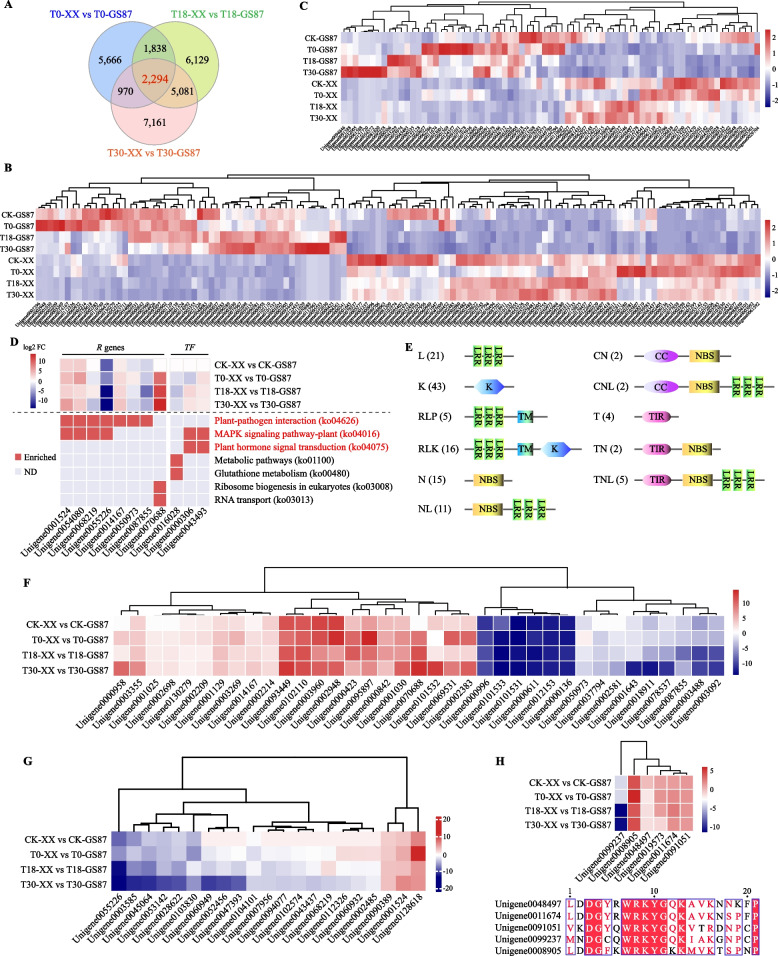
Fig. 4.
Analysis of the 2,294 DEGs shared by GS87 and XX comparisons at 0, 18, and 30 hpi. A Venn diagram showing the numbers of DEGs from GS87 and XX comparisons at 0, 18, and 30 hpi. XX samples were used as controls at each time point. B and C Expression heatmaps of the shared DEGs encoding 126 R proteins and 73 TFs. D Expression heatmap of 11 DEGs enriched in seven KEGG pathways. ND, not detectable. E Schematic structure of 126 R proteins within 11 families based on the disease resistance-related domain type. L, leucine-rich repeat receptor (LRR); K, kinase; RLP, receptor-like protein with transmembrane (TM) domain; RLK, receptor-like kinase with TM domain; N, nucleotide binding site (NBS); NL, NBS-LRR; CN, coiled-coil (CC)-NBS; CNL, CC-NBS-LRR; T, Toll/interleukin receptor (TIR); TN, TIR-NBS; TNL, TIR-NBS-LRR. F and G Clustering analysis of 37 R genes containing the NBS domain and 21 PRR (including RLP and RLK) genes based on log2FC values. H Clustering analysis of six WRKY genes within TF family log2FC values (top) and multiple amino acid sequence alignments of the core domain in the six WRKY proteins (bottom)