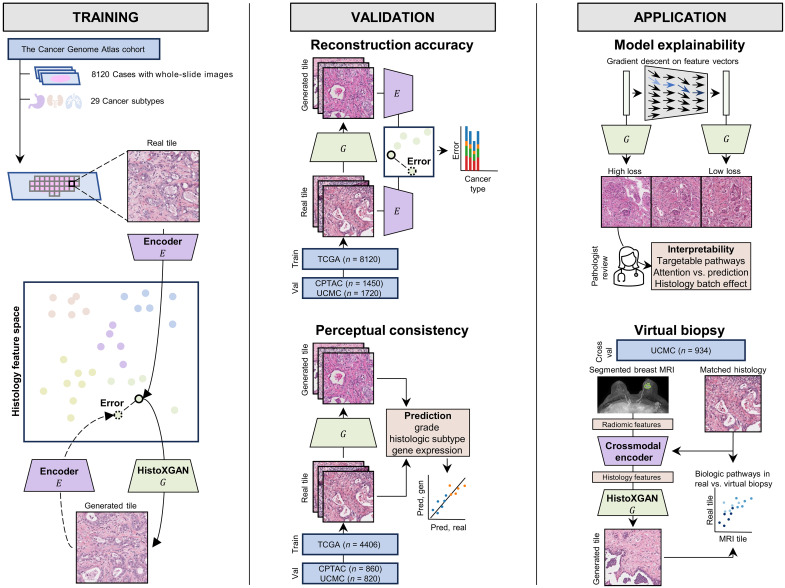
Fig. 1. Overview of HistoXGAN training, validation, and application.
As illustrated on the (left), the HistoXGAN generator G was trained using 8120 cases across 29 cancer types in TCGA. HistoXGAN takes as an input a histologic feature vector derived from any self-supervised feature extractor E and generates a histology tile with near-identical features with respect to the same feature extractor. As shown in the (middle), in this study, we demonstrate that this architecture accurately reconstructs the encoded features from multiple feature extractors in both the training TCGA dataset and external datasets of 3201 slides from 1450 cases from CPTAC and 2656 slides from 1720 cases from University of Chicago Medical Center (UCMC). In addition, we demonstrate that the real and reconstructed images carry near-identical information of interpretable pathologic features such as grade, histologic subtype, and gene expression data. As illustrated to the (right), we showcase the applications of this architecture for model interpretability using gradient descent to illustrate features used in deep learning model predictions. Through systematic review of these features with expert pathologists, we identify characteristics of cancers with targetable pathways, such as homologous recombination deficiency (HRD) and PIK3CA in breast cancer, and illustrate application for attention based models. Last, we train a crossmodal encoder to translate MRI radiomic features into histology features using paired breast MRIs and histology from 934 breast cancer cases from the UCMC and apply HistoXGAN to generate representative histology directly from MRI.