Fig. 2. Reconstruction accuracy in training and validation datasets for CTransPath encoders.
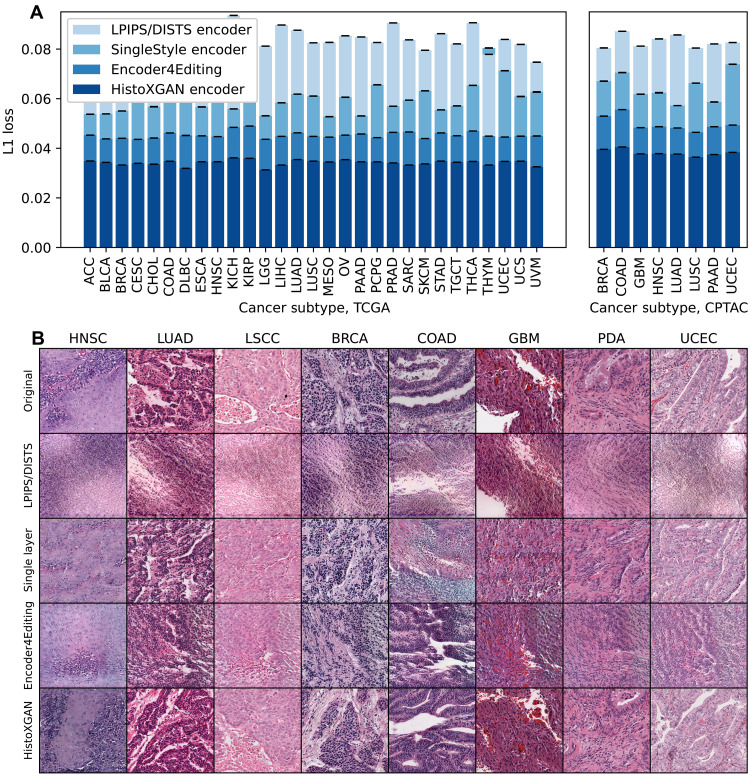
We compare reconstruction accuracy from the real and reconstructed images for HistoXGAN and other architectures for embedding images in GAN latent space. For comparison, we use encoders designed to recreate images from a StyleGAN2 model trained identically to the HistoXGAN model. The Learned Perceptual Image Patch Similarity (LPIPS)/Deep-Image Structure and Texture Similarity (DISTS) encoder uses an equal ratio of LPIPS/DISTS loss between the real and reconstructed images to train the encoder. The Single-Layer and Encoder4Editing encoders are trained to minimize L1 loss between CTransPath feature vector of the real and reconstructed images. (A) HistoXGAN provides more accurate reconstruction of CTransPath features across the TCGA dataset used for GAN training (n = 8120) and solid tumor CPTAC validation (n = 1328) dataset, achieving an average of 30% improvement in L1 loss over the Encoder4Editing encodings in the validation dataset. (B) HistoXGAN reconstructed images consistently provided more accurate representations of features from the input image across cancer types in the CPTAC validation dataset.