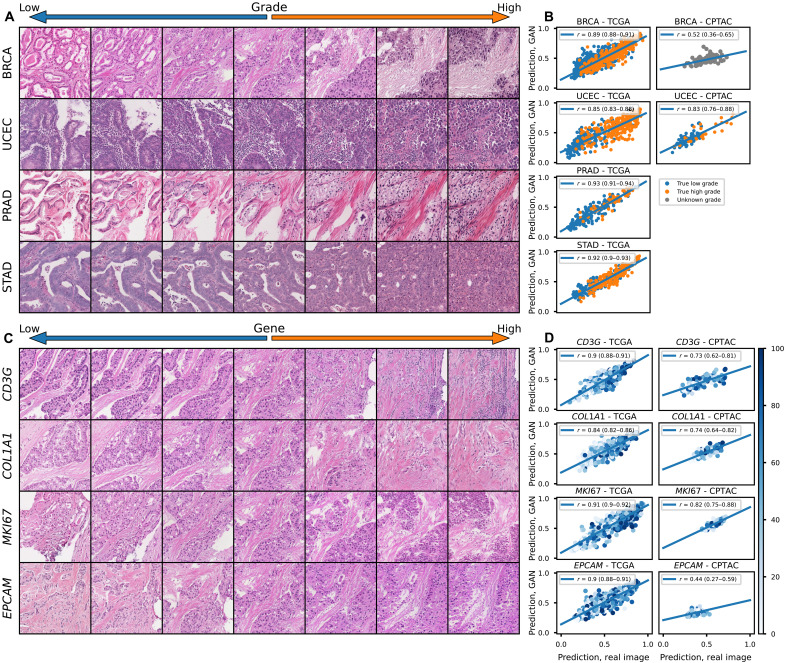
Fig. 3. Perceptual consistency of tumor grade and gene expression in reconstructed images.
(A) Illustration of transition between low and high grade (defined as grade 3 for breast, uterine, or stomach, and Gleason grades 9 or 10 for prostate) across a single image from four cancer types. A vector representing high grade is derived from the coefficients of a logistic regression predicting grade from CTransPath features. This vector is subtracted from the base image to visualize lower grade and added to the base image to visualize higher grade. (B) Correlation between predictions of grade from real and reconstructed tiles, averaged per patient, across cancer types, demonstrating a high perceptual similarity of the grade of the real and generated images. For the TCGA datasets, a deep learning model was trained to predict grade from real tiles for each cancer type using threefold cross-validation. The correlation between predictions for real/generated images is aggregated for the three held-out validation sets. For the CPTAC validation, a deep learning model trained across the entire corresponding TCGA dataset was used to generate predictions. True pathologist-determined high versus low grade is annotated on the images when available. (C) Illustration of transition between expression of select genes across a single image from TCGA-BRCA. (D) Correlation between predictions of gene expression from real and reconstructed tiles, averaged per patient, demonstrating a high perceptual similarity of the gene expression of the real and generated images. True gene expression as a percentile from 0 to 100 is indicated by the color of each data point.