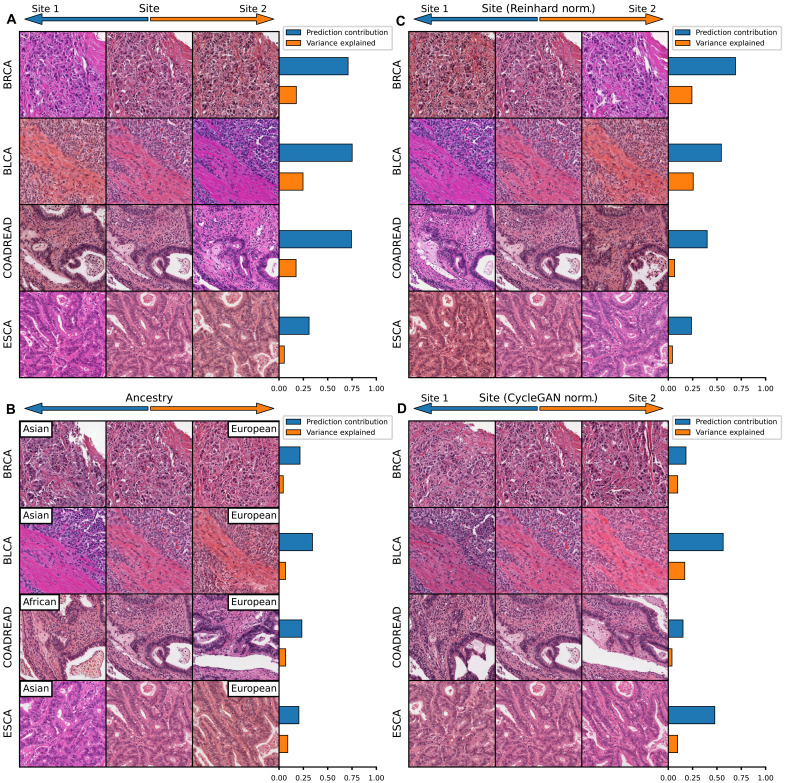
Fig. 4. Visualizing histology batch effect and mitigation with normalization.
Tile-based weakly supervised models were trained to predict tissue source site and patient ancestry class (a batch confounded outcome) across select cancer subtypes in TCGA. The gradient with respect to a prediction of these outcomes was calculated for the average feature vector across each slide in the dataset. Principal components analysis was applied to these gradients, and components were sorted by the magnitude of difference of the component between gradients toward each outcome class. The results are then illustrated for this first principal component (i.e., the component contributing most to model prediction). (A) Model predictions for source site are highly homogenous, with an average 54% of the difference in gradients due to the first principal component. Perturbation of images along this component illustrate that it largely represents change in the staining pattern of slides. (B) Slide stain patterns also contribute to prediction of ancestry, although this first principal component constitutes a smaller proportion of gradients. (C) Reinhard normalization does not eliminate the impact of stain pattern on prediction of site, although it leads to an inversion of the stain detected by the model, perhaps due to overcorrection during normalization. (D) CycleGAN normalization reduces the dependence of predictions on a single principal component, and this most predictive component is no longer clearly indicative of staining differences.