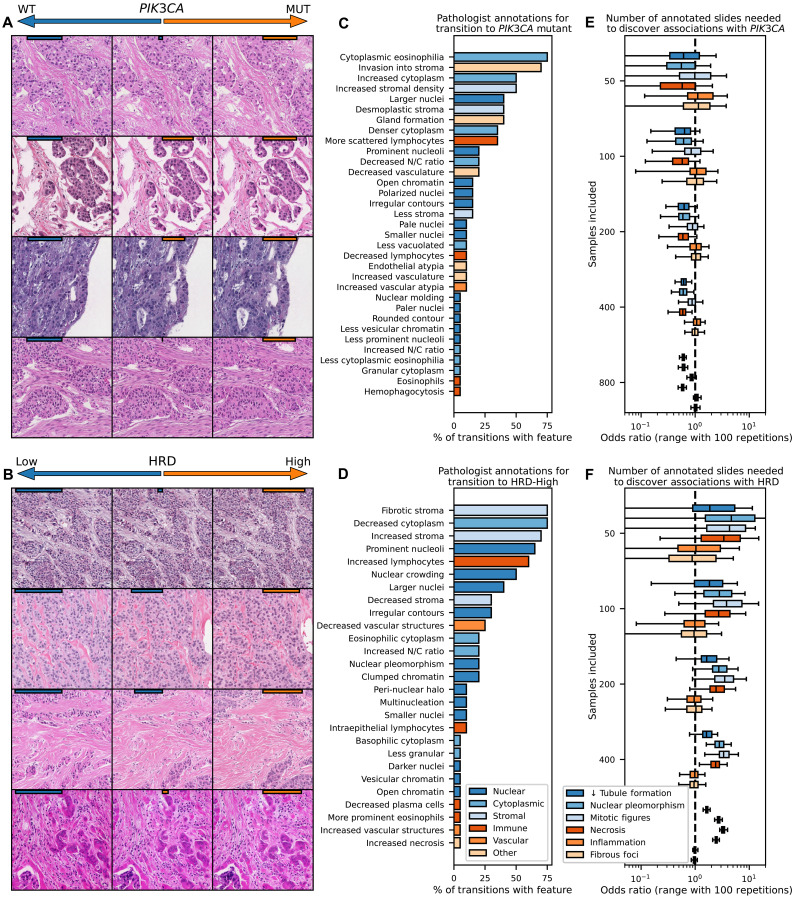
Fig. 5. Illustration of model predictions for targetable alterations in breast cancer.
(A and B) Models were trained to predict PIK3CA mutations and HRD across the TCGA-BRCA dataset (n = 963 for PIK3CA, n = 871 for HRD), as these pathways are common and have Food and Drug Administration–approved therapies. Gradient descent was used to adjust images to maximize/minimize model prediction of PIK3CA/HRD status (with model prediction strength illustrated with orange/blue bars on top of images). Transition to PIK3CA alteration was morphologically associated with increase in abundance and eosinophilic appearance of cytoplasm and stroma, increased tubule formation, and decreased nuclear to cytoplasmic ratio. Transition to high HRD score was associated with nuclear crowding and pleomorphism with occasional multinucleated cells, an increased nuclear/cytoplasmic ratio, increased lymphocytosis, and tumor cell necrosis. (C and D) Structured pathologist review of 20 transitions from low to high model prediction highlight features associated with the selected genomic alterations. (E and F) To determine how many histology slides would be needed to be annotated through traditional methods to uncover these same associations, adjusted odds ratio (estimated through 100 iterations of sampling of listed number of slides) of the association of histologic features with PIK3CA and HRD status are shown as a function of available slides. Tubule formation in PIK3CA and tumor necrosis in HRD that were evident on review of 20 image transitions would require annotation of 400 slides with traditional histologic review to uncover significant associations.