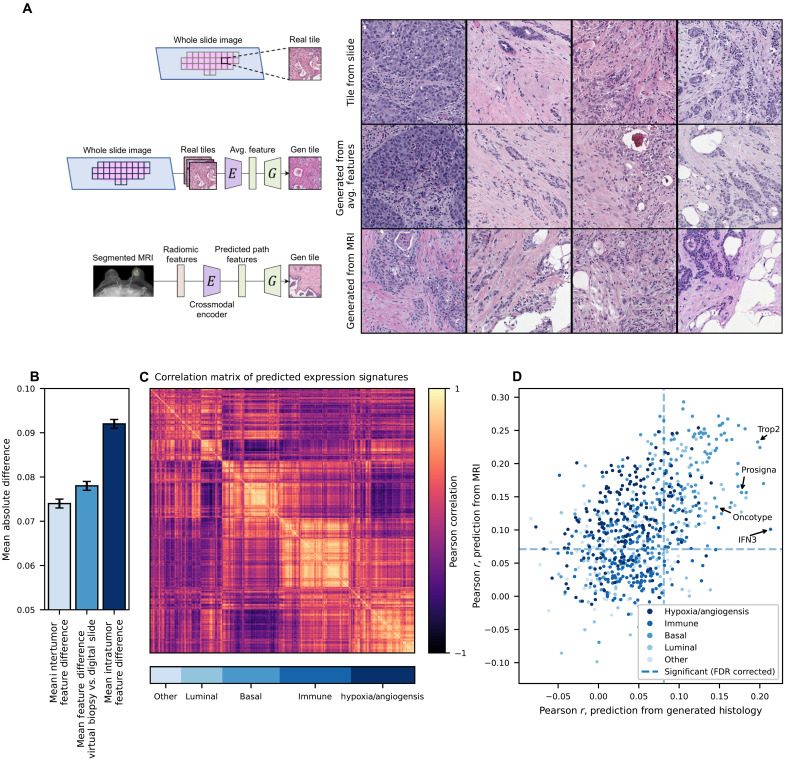
Fig. 6. A virtual biopsy reconstructing histology from radiomic features.
An encoder was trained to predict a slide-level average SSL histology feature vector, using 16,379 radiomic features extracted systematically from 934 breast tumors with paired MRI and digital histology available. Fivefold cross-validation was performed, with predictions pooled from the held-out test set. (A) Representative images from the scanned histology slide, reconstruction of the image from an average SSL histology feature vector, and reconstruction directly from radiomic features (predictions made using cases from the test set for each fold). (B) The mean difference between slide-level average features and features from MRI virtual biopsy reconstruction (middle column) was close to the mean difference between tile features within tumors (left) and much less than the mean difference of image tile features between tumors (right). (C) To explore the biologic accuracy of generated images, we used 775 models trained in TCGA to predict RNA signatures as well as models to predict pathologist annotations of grade and histologic subtype. Predictions from these models largely fell into five orthogonal categories as shown in a correlation matrix. (D) Accuracy of predictions for these 777 RNA/histologic features were similar from generated histology and directly from MRI radiomics (without the intermediary of generated histology); in other words, features could only be predicted from generated histology if they were predictable from MRI radiomics. A number of clinically relevant prognostic signatures such as Prosigna and Oncotype were predictable from generated histology.