FIGURE 1.
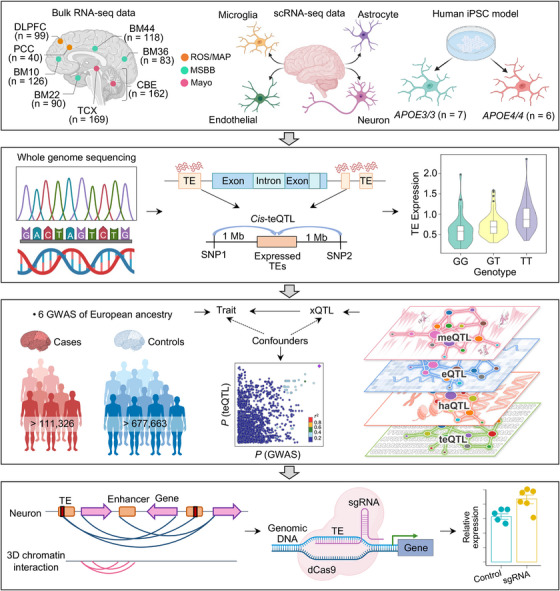
A diagram illustrating systematic characterization of TE dysregulation in human brains with AD. Bulk RNA‐seq data sets from three brain biobanks (MSBB, Mayo, and ROS/MAP) were downloaded from the AD knowledge portal. RNA‐seq data from the MSBB cohort was derived from four brain BM, including BM10, BM22, BM36, and BM44. RNA‐seq data from the Mayo cohort consisted of 156 AD patients and 175 cognitive HCs from cerebellum and TCX tissues. RNA‐seq data from the ROS/MAP cohort were collected from 98 AD and 41 cognitive HCs in two cortex regions. Results obtained from bulk RNA‐seq data were validated using three brain cell‐type RNA‐seq datasets: (1) the first FACS‐purified RNA‐seq data from four major brain cell populations from AD and control frozen cortex tissues, (2) the second snRNA‐seq datasets consisting of 482,472 nuclei from non‐demented control brains and AD brains with both Aβ and tau pathology, and (3) the third RNA‐seq data from human iPSC‐derived population APOE brain microglia cells. Family‐ and locus‐based TE expression levels were calculated using three tools (see Methods). We profiled Atlas of Human Brain teQTLs by integrating locus‐based expressed TEs with corresponding WGS data from the Mayo brain biobank. We then leveraged colocalization analysis of xQTLs with three AD GWAS summary statistics data to identify AD likely causal genes regulated by brain teQTLs. The underlying mechanisms between TE and AD causal genes were dissected using brain cell type–specific enhancer–promoter interactome maps. Finally, we used CRISPRi to confirm the regulatory relationship between locus TE and its target gene. Aβ, amyloid beta; AD, Alzheimer's disease; APOE, apolipoprotein E; BM, Brodmann areas; CRISPRi, CRISPR interference; DLPFC, dorsolateral prefrontal cortex; FACS, fluorescence‐activated cell sorting; GWAS, genome‐wide association study; HC, healthy controls; iPSC, induced pluripotent stem cell; Mayo, Mayo Clinic; MSBB, Mount Sinai brain bank; PCC, posterior cingulate cortex; RNA‐seq, RNA sequencing; ROS/MAP, Religious Orders Study/Rush Memory and Aging Project; TCX, temporal cortex; TE, transposable element; teQTLs, transposable element quantitative trait loci; WGS, whole‐genome sequencing.
