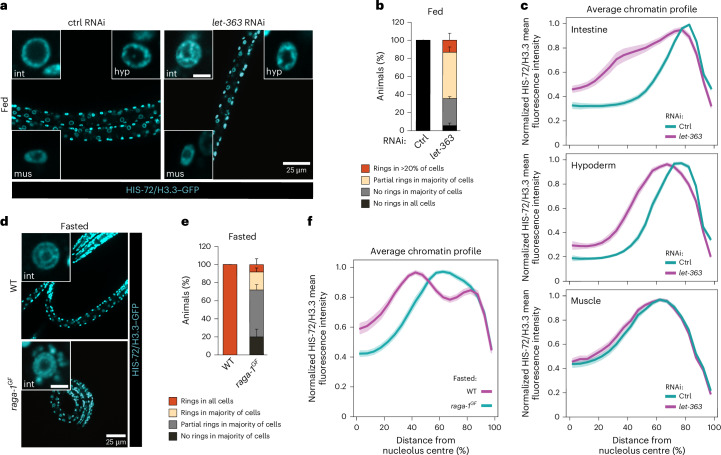
Fig. 3. mTORC1 signalling is necessary and sufficient to regulate 3D genome architecture in response to nutrients in intestine.
a, Single focal planes of representative fed L1 expressing HIS-72/H3.3–GFP upon control (ctrl) or let-363 RNAi treatment. Insets: zoom of single nucleus of the indicated tissue. Hyp, hypoderm; int, intestine; mus, muscle. Inset scale bar, 2.5 μm. b, Quantification of the percentage of fed animals under control or let-363 RNAi, within the indicated categories for 3D chromatin organization in intestine. Data are shown as mean ± s.e.m. of three independent biological replicas. c, Line plots of the averaged single nuclei profile of HIS-72/H3.3–GFP from 72 intestinal, hypodermal or muscle nuclei of fed animals upon control and let-363 RNAi, showing the radial fluorescence intensity as a function of the relative distance from the nucleolus centre. Data are from three independent biological replicas. For let-363 RNAi, larvae in proportions to their relative abundance within the chromatin organization categories as in b, were analysed for all tissues. The shaded area represents the 95% confidence interval of the mean profile. Profiles with a single peak were compared to estimate the statistical significance of differences, as described in Methods. P values are given in Supplementary Table 2. d, As in a but showing fasted WT and raga-1GF animals expressing HIS-73/H3.3–GFP. Insets: zoom of single nucleus of the intestine. e, Quantification of the percentage of fasted WT and raga-1GF animals in the indicated categories for 3D chromatin organization in intestine. Data are shown as mean ± s.e.m. of three independent biological replicas. f, Line plots as in c but of the averaged single nuclei profile of HIS-72/H3.3–GFP from 72 intestinal nuclei of fasted WT and raga-1GF larvae. Data are from three and four independent biological replicas for raga-1GF and WT, respectively. For raga-1GF, larvae in proportions to their relative abundance within the chromatin organization categories as in e, were analysed. For c and f, heat maps of single nuclei profiles are provided in Extended Data Fig. 3h,p, respectively. Source numerical data are available in Source data.