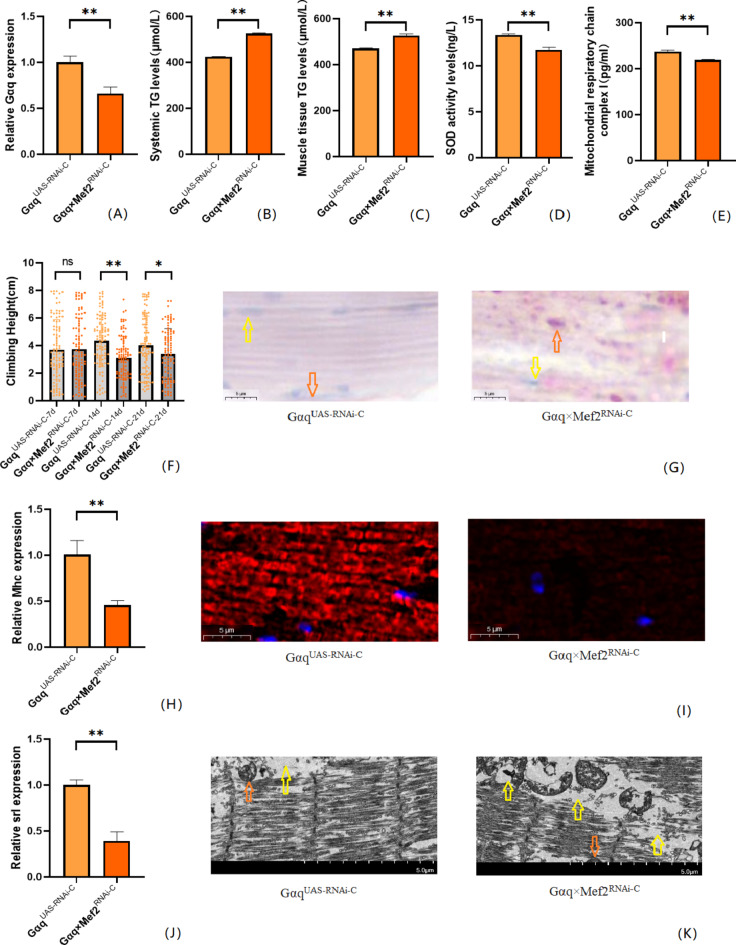
Fig. 4.
Relative gene expression, lipid metabolism levels, oxidative capacity, mitochondrial function, and 3 s climbing ability in skeletal muscle tissue Gαq knockdown of Drosophila skeletal muscle. (A) Relative expression of Gαq, Sample n = 50. (B) Whole Body Triglyceride Levels, Sample n = 20. (C) Muscle Tissue Triglyceride Levels, Sample n = 30. (D) SOD activity levels, Sample n = 30. (E) Mitochondrial respiratory chain complex I content, Sample n = 30. (F) Climbing height, Sample n = 100. (G) Oil red staining, Sample n = 5. Yellow arrows indicate nuclei and orange arrows indicate lipid accumulation (scale: black line is 5 μm). (H) Relative expression of Mhc, Sample n = 50. (I) Immunofluorescence image of myosin heavy chain, Sample n = 5, blue dots are nuclei and red fluorescent bands are myogenic fibers (scale: white line is 5 μm). (J) Relative expression of Srl, Sample n = 50. (K) Transmission electron micrograph of skeletal muscle, Sample n = 5, orange indicates nuclei, yellow indicates muscle fiber damage (scale, white line is 5 μm). The one-way analysis of variance (ANOVA) with least significant difference (LSD) tests was used to identify difference among the groups. Data are represented as mean ± standard deviation. *P < 0.05; **P < 0.01; n s means no significant difference.